Abstract
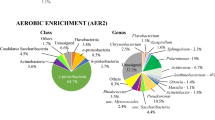
Petroleum and fuel oil are complex mixtures of recalcitrant hydrocarbons. The biodegradation of these hydrocarbons needs the action of a vast variety of enzymatic capacities. A microbial consortium offers the capability to degrade complex substrates through the assembly of different biochemical reactions, providing a metabolic versatility superior to axenic cultures. In this work, the microbial population dynamics, taxonomy, and the catabolic capacity of a stabilized consortium exposed to fuel and crude oil was analyzed through metagenomics. The stabilized consortium degraded 59% of crude oil components after 8 days, and 34% of fuel oil components after 130 days. Population dynamics analysis indicates that in fuel oil the biodiversity richness was higher; however, denaturing gradient gel electrophoresis similarity dendrogram shows significant changes in the microbial population during crude oil degradation. Taxonomy studies indicate a great genera divergence; only eight microbial genera were common in both samples. In crude oil, the Limnobacter sp. was the most abundant specie (15.6%), while Sphingomonas wittichii (7.9%) and Novosphingobium aromaticivorans (7.6%) were abundant in fuel oil. These microorganisms have been reported to participate in the degradation of aliphatic and aromatic hydrocarbons. Functional analysis suggests that fuel and crude oil components changed the interactions between the consortium members affecting the collective metabolic functionality.







Similar content being viewed by others
References
Balachandran C, Duraipandiyan V, Balakrishna K, Ignacimuthu S (2012) Petroleum and polycyclic aromatic hydrocarbons (PAHs) degradation and naphthalene metabolism in Streptomyces sp. (ERI-CPDA-1) isolated from oil contaminated soil. Bioresour Technol 112:83–90. doi:10.1016/j.biortech.2012.02.059
Clarke KR, Warwick RM (2001) Change in marine communities: an approach to statistical analysis and interpretation, 2nd edn. Primer-E, Plymouth
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi:10.1093/nar/gkn879
Diggle SP, Gardner A, West SA, Griffin AS (2007) Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Philos Trans R Soc B Biol Sci 362:1241. doi:10.1098/rstb.2007.2049
Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD (2008) Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. doi:10.1186/1471-2180-8-43
Fang H, Cai L, Yu Y, Zhang T (2013) Metagenomic analysis reveals the prevalence of biodegradation genes for organic pollutants in activated sludge. Bioresour Technol 129:209–218. doi:10.1016/j.biortech.2012.11.054
Gontcharova V, Youn E, Wolcott RD, Hollister EB, Gentry TJ, Dowd SE (2010) Black box chimera check (B2C2): a windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol J 4:47–52. doi:10.2174/1874285801004010047
Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, Estrada DA, Dowd SE, Mueller UG (2011) Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb Ecol 61:821–831. doi:10.1007/s00248-010-9793-4
Joshi M, Dhebar S, Dhebar S, Bhargava P, Pandit A, Patel R, Saxena A, Bagatharia S (2014) Metagenomics of petroleum muck: revealing microbial diversity and depicting microbial syntrophy. Arch Microbiol 196:531–544
Kostichka K, Thomas SM, Gibson KJ, Nagarajan V, Cheng Q (2001) Cloning and characterization of a gene cluster for cyclododecanone oxidation in Rhodococcus ruber SC1. J Bacteriol 183:6478–6486. doi:10.1128/JB.183.21.6478-6486.2001
Kumar DK, Kumar P, Kumar SP, Shukla P (2014) Exploring prospects of monooxygenase-based biocatalysis in xenobiotics. In: Microbial biodegradation and bioremediation. Elsevier, Oxford, pp 577–614. doi:10.1016/B978-0-12-800021-2.00026-1
Kuppusamy S, Thavamani P, Megharaj M, Naidu R (2016) Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by novel bacterial consortia tolerant to diverse physical settings—assessments in liquid- and slurry-phase systems. Int Biodeterior Biodegrad 108:149–157. doi:10.1016/j.ibiod.2015.12.013
Liu B, Ju M, Liu J, Wu W, Li X (2016) Isolation, identification, and crude oil degradation characteristics of a high-temperature, hydrocarbon-degrading strain. Mar Pollut Bull 106:301–307. doi:10.1016/j.marpolbul.2015.09.053
McCormick MS, Lippard SJ (2011) Analysis of substrate access to active sites in bacterial multicomponent monooxygenase hydroxylases: X-ray crystal structure of xenon-pressurized phenol hydroxylase from Pseudomonas sp. OX1. Biochemstry 51:11058–11069. doi:10.1021/bi201248b
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform 9:386. doi:10.1186/1471-2105-9-386
Murrell CJ, Gilbert B, McDonald IR (2000) Molecular biology and regulation of methane monooxygenase. Arch Microbiol 173:325–332. doi:10.1007/s002030000158
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Parks DH, Beiko RG (2015) STAMP: statistical analysis of metagenomic profiles. In: Nelson KE (ed) Encyclopedia of metagenomics: genes, genomes and metagenomes: basics, methods, databases and tools. pp 641–645. doi: 10.1007/978-1-4899-7478-5_780
Pi Y, Meng L, Bao M, Sun P, Lu J (2016) Degradation of crude oil and relationship with bacteria and enzymatic activities in laboratory testing. Int Biodeterior Biodegrad 106:106–116. doi:10.1016/j.ibiod.2015.10.015
Qu Y, Shi S, Zhou H, Ma Q, Li X, Zhang X, Zhou J (2012) Characterization of a novel phenol hydroxylase in indoles biotransformation from a strain Arthrobacter sp. W1. PLoS ONE 7:e44313. doi:10.1371/journal.pone.0044313
Rojas-Herrera R, Narváez-Zapata J, Zamudio-Maya M, Mena-Martínez M (2008) A Simple silica-based method for metagenomic DNA extraction from soil and sediments. Mol Biotechnol 40:13–17. doi:10.1007/s12033-008-9061-8
Salamanca D, Engesser KH (2014) Isolation and characterization of two novel strains capable of using cyclohexane as carbon source. Environ Sci Pollut Res 21:12757–12766. doi:10.1007/s11356-014-3206-z
Seeger M, Cámara B, Hofer B (2001) Dehalogenation, denitration, dehydroxylation, and angular attack on substituted biphenyls and related compounds by a biphenyl dioxygenase. J Bacteriol 183:3548–3555. doi:10.1128/JB.183.12.3548-3555.2001
Speight JG (2006) The chemistry and technology of petroleum, 4th edn. CRC Press, Boca Raton
Tan CH, Koh KS, Xie C, Zhang J, Tan XH, Lee GP, Zhou Y, Ng WJ, Rice SA, Kjelleberg S (2015) Community quorum sensing signalling and quenching: microbial granular biofilm assembly. NPJ Biofilms Microbiomes 1:15006. doi:10.1038/npjbiofilms.2015.6
Thavamani P, Malik S, Beer M, Megharaj M, Naidu R (2012) Microbial activity and diversity in long-term mixed contaminated soils with respect to polyaromatic hydrocarbons and heavy metals. J Environ Manag 99:10–17. doi:10.1016/j.jenvman.2011.12.030
Vedler E, Heinaru E, Jutkina J, Viggor S, Koressaar T, Remm M, Heinaru A (2013) Limnobacter spp. as newly detected phenol-degraders among Baltic Sea surface water bacteria characterised by comparative analysis of catabolic genes. Syst Appl Microbiol 8:525–532. doi:10.1016/j.syapm.2013.07.004
Viggor S, Juhanson J, Jöesaar M, Mitt M, Truu J, Vedler E, Heinaru A (2013) Dynamic changes in the structure of microbial communities in Baltic Sea coastal seawater microcosms modified by crude oil, shale oil or diesel fuel. Microbiol Res 168:415–427. doi:10.1016/j.micres.2013.02.006
Yergeau E, Sanschagrin S, Beaumier D, Greer CW (2012) Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high arctic soils. PLoS ONE 7:e30058. doi:10.1371/journal.pone.0030058
Acknowledgements
M. Canul-Chan gratefully acknowledges the scholarship (No. 220532) from CONACyT to pursue his graduate studies.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editorial responsibility: M. Abbaspour.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Canul-Chan, M., Sánchez-González, M., González-Burgos, A. et al. Population structures shift during the biodegradation of crude and fuel oil by an indigenous consortium. Int. J. Environ. Sci. Technol. 15, 1–16 (2018). https://doi.org/10.1007/s13762-017-1362-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-017-1362-7