Abstract
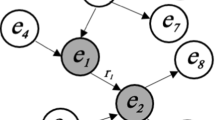
Diagnosis prediction, a key factor in enhancing healthcare efficiency, remains a focal point in clinical decision support research. However, the time-series, sparse and multi-noise characteristics of electronic health record (EHR) data make it a great challenge. Existing methods commonly address these issues using RNNs and incorporating medical prior knowledge from medical knowledge bases, but they neglect the local spatial characteristics and spatial–temporal correlation of the data. Consequently, we propose MDPG, a diagnosis prediction model based on patient knowledge graphs. Initially, we represent the electronic visit records of patients as a patient-centered temporal knowledge graph, capturing the local spatial structure and temporal characteristics of the visit information. Subsequently, we design the spatial graph convolution block, temporal self-attention block, and spatial–temporal synchronous graph convolution block to capture the spatial, temporal, and spatial–temporal correlations embedded in them, respectively. Ultimately, we accomplish the prediction of patients’ future states through multi-label classification. We conduct comprehensive experiments on two real-world datasets independently and evaluate the results using visit-level precision@k and code-level accuracy@k metrics. The experimental results demonstrate that MDPG outperforms all baseline models, yielding the best performance.





Similar content being viewed by others
Data availability
The MIMIC-III dataset are publicly available at https://mimic.mit.edu/. The MedClin dataset is not currently available as a private dataset.
References
Birkhead GS, Klompas M, Shah NR. Uses of electronic health records for public health surveillance to advance public health. Annu Rev Public Health. 2015;36:345–59.
Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13(6):395–405.
Choi E, Bahadori MT, Searles E, Coffey C, Thompson M, Bost J, Tejedor-Sojo J, Sun J. Multi-layer representation learning for medical concepts. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, 2016;1495–1504.
Zhou J, Sun J, Liu Y, Hu J, Ye J. Patient risk prediction model via top-k stability selection. In: Proceedings of the 2013 SIAM international conference on data mining, 2013; 55–63. SIAM.
Choi E, Bahadori MT, Sun J, Kulas J, Schuetz A, Stewart W. Retain: An interpretable predictive model for healthcare using reverse time attention mechanism. Adv Neural Inf Process Syst. 2016;29:3512–20.
Weston J, Chopra S, Bordes A. Memory networks. In: 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7–9, 2015, Conference Track Proceedings.
Song H, Rajan D, Thiagarajan J, Spanias A. Attend and diagnose: Clinical time series analysis using attention models. In: Proceedings of the AAAI Conference on Artificial Intelligence, 2018; vol. 32.
Ma F, Chitta R, Zhou J, You Q, Sun T, Gao J. Dipole: Diagnosis prediction in healthcare via attention-based bidirectional recurrent neural networks. In: Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, 2017; pp. 1903–1911.
Schuster M, Paliwal KK. Bidirectional recurrent neural networks. IEEE Trans Signal Process. 1997;45(11):2673–81.
Wei W-Q, Bastarache LA, Carroll RJ, Marlo JE, Osterman TJ, Gamazon ER, Cox NJ, Roden DM, Denny JC. Evaluating phecodes, clinical classification software, and ICD-9-cm codes for phenome-wide association studies in the electronic health record. PLoS ONE. 2017;12(7):0175508.
Stearns MQ, Price C, Spackman KA, Wang AY. Snomed clinical terms: overview of the development process and project status. In: Proceedings of the AMIA Symposium, 2001; p. 662
Choi E, Bahadori MT, Song L, Stewart WF, Sun J. Gram: graph-based attention model for healthcare representation learning. In: Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, 2017; pp. 787–795.
Ma F, You Q, Xiao H, Chitta R, Zhou J, Gao J. Kame: Knowledge-based attention model for diagnosis prediction in healthcare. In: Proceedings of the 27th ACM international conference on information and knowledge management, 2018; pp. 743–752.
Gao J, Wang X, Wang Y, Yang Z, Gao J, Wang J, Tang W, Xie X. Camp: co-attention memory networks for diagnosis prediction in healthcare. In: 2019 IEEE international conference on data mining (ICDM), 2019; pp. 1036–1041. IEEE.
Li Y, Qian B, Zhang X, Liu H. Graph neural network-based diagnosis prediction. Big Data. 2020;8(5):379–90.
Kipf TN, Welling M. Semi-supervised classification with graph convolutional networks. In: 5th international conference on learning representations, ICLR 2017, Toulon, France, April 24–26, 2017, Conference Track Proceedings.
Velickovic P, Cucurull G, Casanova A, Romero A, Liò P, Bengio Y. Graph attention networks. In: 6th international conference on learning representations, ICLR 2018, Vancouver, BC, Canada, April 30–May 3, 2018, Conference Track Proceedings.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. Adv Neural Inf Process Syst. 2017;30:1–11.
Ji S, Pan S, Cambria E, Marttinen P, Philip SY. A survey on knowledge graphs: representation, acquisition, and applications. IEEE Trans Neural Netw Learn Syst. 2021;33(2):494–514.
Sankar A, Wu Y, Gou L, Zhang W, Yang H. Dysat: Deep neural representation learning on dynamic graphs via self-attention networks. In: WSDM ’20: The Thirteenth ACM international conference on web search and data mining, Houston, TX, USA, February 3–7, 2020, pp. 519–527.
Song C, Lin Y, Guo S, Wan H. Spatial–temporal synchronous graph convolutional networks: a new framework for spatial–temporal network data forecasting. In: Proceedings of the AAAI conference on artificial intelligence, 2020; vol. 34, pp. 914–921.
Choi E, Du N, Chen R, Song L, Sun J. Constructing disease network and temporal progression model via context-sensitive Hawkes process. In: 2015 IEEE international conference on data mining, 2015; pp. 721–726. IEEE.
Wang X, Sontag D, Wang F. Unsupervised learning of disease progression models. In: Proceedings of the 20th ACM SIGKDD international conference on knowledge discovery and data mining, 2014; pp. 85–94.
Xiao H, Gao J, Vu L, Turaga DS. Learning temporal state of diabetes patients via combining behavioral and demographic data. In: Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, 2017; pp. 2081–2089.
Zhou J, Yuan L, Liu J, Ye J. A multi-task learning formulation for predicting disease progression. In: Proceedings of the 17th ACM SIGKDD international conference on knowledge discovery and data mining, 2011; pp. 814–822.
Che Z, Kale D, Li W, Bahadori MT, Liu Y. Deep computational phenotyping. In: Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining, 2015; pp. 507–516.
Liu C, Wang F, Hu J, Xiong H. Temporal phenotyping from longitudinal electronic health records: a graph based framework. In: Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining, 2015; pp. 705–714.
Zhou J, Wang F, Hu J, Ye J. From micro to macro: data driven phenotyping by densification of longitudinal electronic medical records. In: Proceedings of the 20th ACM sigkdd international conference on knowledge discovery and data mining, 2014; pp. 135–144.
Che Z, Purushotham S, Cho K, Sontag D, Liu Y. Recurrent neural networks for multivariate time series with missing values. Sci Rep. 2018;8(1):6085.
Lipton ZC, Kale DC, Wetzel R, et al. Modeling missing data in clinical time series with RNNS. Mach Learn Healthcare. 2016;56:253–70.
Nguyen P, Tran T, Wickramasinghe N, Venkatesh S. Deepr: a convolutional net for medical records. IEEE J Biomed Health Inf. 2017;21(1):22–30.
Cheng Y, Wang F, Zhang P, Hu J. Risk prediction with electronic health records: a deep learning approach. In: Proceedings of the 2016 SIAM international conference on data mining, 2016; pp. 432–440. SIAM.
Lee W, Park S, Joo W, Moon I-C. Diagnosis prediction via medical context attention networks using deep generative modeling. In: 2018 IEEE international conference on data mining (ICDM), 2018; pp. 1104–1109. IEEE.
Miller AH, Fisch A, Dodge J, Karimi A, Bordes A, Weston J. Key-value memory networks for directly reading documents. In: Proceedings of the 2016 conference on empirical methods in natural language processing, EMNLP 2016, Austin, Texas, USA, November 1–4, 2016, pp. 1400–1409.
Yan S, Xiong Y, Lin D. Spatial temporal graph convolutional networks for skeleton-based action recognition. In: Proceedings of the AAAI conference on artificial intelligence, 2018; vol. 32.
Liang K, Meng L, Liu M, Liu Y, Tu W, Wang S, Zhou S, Liu X, Sun F. Reasoning over different types of knowledge graphs: static, temporal and multi-modal. arXiv:2212.05767
Leblay J, Chekol MW. Deriving validity time in knowledge graph. In: Companion proceedings of the the web conference 2018; pp. 1771–1776
Dasgupta SS, Ray SN, Talukdar PP. Hyte: Hyperplane-based temporally aware knowledge graph embedding. In: EMNLP, 2018; pp. 2001–2011
García-Durán A, Dumancic S, Niepert M. Learning sequence encoders for temporal knowledge graph completion. In: Proceedings of the 2018 conference on empirical methods in natural language processing, Brussels, Belgium, October 31–November 4, 2018, pp. 4816–4821.
Leblay J, Chekol MW, Liu X. Towards temporal knowledge graph embeddings with arbitrary time precision. In: Proceedings of the 29th ACM international conference on information & knowledge management, 2020; pp. 685–694.
Nayyeri M, Vahdati S, Khan MT, Alam MM, Wenige L, Behrend A, Lehmann J. Dihedron algebraic embeddings for spatio-temporal knowledge graph completion. In: The Semantic Web—19th international conference, ESWC 2022, Hersonissos, Crete, Greece, May 29–June 2, 2022, Proceedings. Lecture Notes in Computer Science, 2022; vol. 13261, pp. 253–269.
Trivedi R, Dai H, Wang Y, Song L. Know-evolve: deep temporal reasoning for dynamic knowledge graphs. In: International conference on machine learning, 2017; pp. 3462–3471. PMLR.
Jin W, Qu M, Jin X, Ren X. Recurrent event network: autoregressive structure inference over temporal knowledge graphs. In: Proceedings of the 2020 conference on empirical methods in natural language processing, EMNLP 2020, Online, November 16–20, 2020, pp. 6669–6683
Li Z, Jin X, Li W, Guan S, Guo J, Shen H, Wang Y, Cheng X. Temporal knowledge graph reasoning based on evolutional representation learning. In: SIGIR ’21: The 44th international ACM SIGIR conference on research and development in information retrieval, virtual event, Canada, July 11–15, 2021, pp. 408–417.
Zhang M, Xia Y, Liu Q, Wu S, Wang L. Learning latent relations for temporal knowledge graph reasoning. In: Proceedings of the 61st annual meeting of the association for computational linguistics (Volume 1: Long Papers), ACL 2023, Toronto, Canada, July 9–14, 2023, pp. 12617–12631.
Vashishth S, Sanyal S, Nitin V, Talukdar PP. Composition-based multi-relational graph convolutional networks. In: 8th international conference on learning representations, ICLR 2020, Addis Ababa, Ethiopia, April 26–30, 2020.
Pareja A, Domeniconi G, Chen J, Ma T, Suzumura T, Kanezashi H, Kaler T, Schardl T, Leiserson C. Evolvegcn: Evolving graph convolutional networks for dynamic graphs. In: Proceedings of the AAAI conference on artificial intelligence, 2020; vol. 34, pp. 5363–5370.
You J, Du T, Leskovec J. ROLAND: graph learning framework for dynamic graphs. In: Zhang, A., Rangwala, H. (eds.) KDD ’22: The 28th ACM SIGKDD conference on knowledge discovery and data mining, Washington, DC, USA, August 14–18, 2022, pp. 2358–2366.
Zhang C, Yao Z, Yao H, Huang F, Chen CLP. Dynamic representation learning via recurrent graph neural networks. IEEE Trans Syst Man Cybern Syst. 2023;53(2):1284–97.
Gehring J, Auli M, Grangier D, Yarats D, Dauphin YN. Convolutional sequence to sequence learning. In: International conference on machine learning, 2017; pp. 1243–1252. PMLR.
Johnson AE, Pollard TJ, Shen L, Lehman L-WH, Feng M, Ghassemi M, Moody B, Szolovits P, AnthonyCeli L, Mark RG. Mimic-iii, a freely accessible critical care database. Sci Data. 2016;3(1):1–9.
Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng C-K, Stanley HE. Physiobank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals. Circulation. 2000;101(23):215–20.
Cho K, Merrienboer B, Gülçehre Ç, Bahdanau D, Bougares F, Schwenk H, Bengio Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, EMNLP 2014, October 25-29, 2014, Doha, Qatar, A Meeting of SIGDAT, a Special Interest Group of The ACL, pp. 1724–1734.
Kingma DP, Ba J. Adam: A method for stochastic optimization. In: 3rd International conference on learning representations, ICLR 2015, San Diego, CA, USA, May 7–9, 2015, Conference Track Proceedings.
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016; pp. 770–778.
Acknowledgements
We would like to thank the anonymous reviewers for their insightful suggestions. Our work is supported by the National Key Research and Development Program of China (Grant No. 2020AAA0109400).
Author information
Authors and Affiliations
Contributions
WW designed the study, performed measurements, designed the analysis, and wrote the manuscript. YF designed the schema for PKG and extracted the data. HZ designed the analysis and the schema for PKG. XW designed the analysis. RC cleaned the clinical data. WC designed the schema for PKG. XZ designed the study and the analysis. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no potential conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, W., Feng, Y., Zhao, H. et al. Mdpg: a novel multi-disease diagnosis prediction method based on patient knowledge graphs. Health Inf Sci Syst 12, 15 (2024). https://doi.org/10.1007/s13755-024-00278-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13755-024-00278-7