Abstract
Objective and Methods
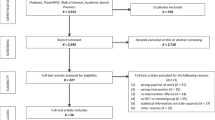
Understanding the role genetics plays in diseases that are acquired as a result of workplace exposures and the interaction between genes and workplace-environmental factors is extremely important. The reason genetic research has become relevant with regard to occupational health is due to the enormous technological advances in molecular biology and genetics in recent years. Literature searches were performed using the many sites including PubMed, Google Scholar, ScienceDirect, and further refined by reviewing titles, abstracts, and references in the bibliography to confirm additional missing case reports.
Results and Conclusion
The purpose of this review is to consolidate the diverse literatures on genetics in the workplace. Focusing on maintaining a healthy workplace, it can change from controlling the environment to excluding vulnerable workers. Testing of genes should only be done with workers’ agree and with the employee controlling access to that information. Although genetic testing has the potential to improve worker health and reduce occupational diseases, there are some concerns regarding clinical relevance, such as autonomy and privacy violations.
Similar content being viewed by others
References
Wright, L. Understanding Genetics: A Primer for Occupational Health Practice. AAOHN J. 53, 534–542 (2005).
Marchant, G. E. Genomics and toxic substances. Part 2: Genetic susceptibility to environmental agents. Environ. Law Rep. 33, 10641–10667 (2003).
Rachiglio, A. M. et al. Limits and potential of targeted sequencing analysis of liquid biopsy in patients with lung and colon carcinoma. Oncotarget 7, 66595–66605 (2016).
Xia, S. et al. Genomic variations in plasma cell free DNA differentiate early stage lung cancers from normal controls. Lung Cancer 90, 78–84 (2015).
Couraud, S. et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin. Cancer Res. 20, 4613–4624 (2014).
Hicks, J. K., Saller, J., Wang, E., Boyle, T. & Gray, J. E. Cell-free circulating tumor DNA supplementing tissue biopsies for identification of targetable mutations: Implications for precision medicine and considerations for reconciling results. Lung Cancer 111, 135–138 (2017).
He, Y. et al. Detection of cancer specific mutations in early-stage non-small cell lung cancer using cell-free DNA by targeted sequencing. Int. J. Oncol. 49, 2351–2358 (2016).
Buttitta, F. et al. Effective assessment of EGFR mutation status in bronchoalveolar lavage and pleural fluids by next-generation sequencing. Clin. Cancer Res. 19, 691–698 (2013).
Marchetti, A. et al. Complex mutations & subpopulations of deletions at exon 19 of EGFR in NSCLC revealed by next generation sequencing: potential clinical implications. PLoS One 7, e42164 (2012).
Que, D. et al. EGFR mutation status in plasma and tumor tissues in non-small cell lung cancer serves as a predictor of response to EGFR-TKI treatment. Cancer Biol. Ther. 17, 320–327 (2016).
Yoon, S. H. et al. Peptide Nucleic Acid Clamping Versus Direct Sequencing for the Detection of EGFR Gene Mutation in Patients with Non-small Cell Lung Cancer. Cancer Res. Treat. 47, 661–669 (2015).
Malapelle, U. et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl. Lung Cancer Res. 5, 505–510 (2016).
Malapelle, U. et al. Consistency and reproducibility of next-generation sequencing and other multigene mutational assays: A worldwide ring trial study on quantitative cytological molecular reference specimens. Cancer Cytopathology 125, 615–626 (2017).
Bartels, S. et al. Molecular Analysis of Circulating Cell-Free DNA from Lung Cancer Patients in Routine Laboratory Practice: A Cross-Platform Comparison of Three Different Molecular Methods for Mutation Detection. J. Mol. Diagn. 19, 722–732 (2017).
Bennett, C. W., Berchem, G., Kim, Y. J. & El-Khoury, V. Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget 7, 71013–71035 (2016).
Oxnard, G. R. et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. 20, 1698–1705 (2014).
Pécuchet, N., Legras, A., Laurent-Puig, P. & Blons, H. Lung cancer molecular testing, what role for Next Generation Sequencing and circulating tumor DNA. Ann. Pathol. 36, 80–93 (2016).
Pécuchet, N. et al. Base-Position Error Rate Analysis of Next-Generation Sequencing Applied to Circulating Tumor DNA in Non-Small Cell Lung Cancer: A Prospective Study. PLoS Med. 13, doi:10.1371/journal. pmed.1002199 (2016).
Daniels, M. et al. Whole genome sequencing for lung cancer. J. Thorac. Dis. 4, 155–163 (2012).
Thompson, J. C. et al. Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin. Cancer Res. 22, 5772–5782 (2016).
Tuononen, K. et al. Comparison of targeted next-generation sequencing (NGS) and real-time PCR in the detection of EGFR, KRAS, and BRAF mutations on formalin-fixed, paraffin-embedded tumor material of nonsmall cell lung carcinoma-superiority of NGS. Genes Chromosomes Cancer 52, 503–511 (2013).
Malapelle, U. et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br. J. Cancer 116, 802–810 (2017).
Takase, Y. et al. Highly sensitive detection of a HER2 12-base pair duplicated insertion mutation in lung cancer using the Eprobe-PCR method. PLoS One 12, doi:10.1371/journal.pone.0171225 (2017).
Rouanne, M. et al. Osteopontin and thrombospondin-1 play opposite roles in promoting tumor aggressiveness of primary resected non-small cell lung cancer. BMC Cancer 16, doi:10.1186/s12885-016-2541-5 (2016).
Meredith, S. L. et al. Irradiation Decreases the Neuroendocrine Biomarker Pro-Opiomelanocortin in Small Cell Lung Cancer Cells In Vitro and In Vivo. PLoS One 11, doi:10.1371/journal.pone.0148404 (2016).
Lou, T. F. et al. Cancer-Specific Production of N-Acetylaspartate via NAT8L Overexpression in Non-Small Cell Lung Cancer and Its Potential as a Circulating Biomarker. Cancer Prev. Res. (Phila) 9, 43–52 (2016).
Lococo, F. et al. Preliminary Evidence on the Diagnostic and Molecular Role of Circulating Soluble EGFR in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 16, 19612–19630 (2015).
Gumireddy, K. et al. AKAP4 is a circulating biomarker for non-small cell lung cancer. Oncotarget 6, 17637–17647 (2015).
Edelman, M. J. et al. GP88 (progranulin): a novel tissue and circulating biomarker for non-small cell lung carcinoma. Hum. Pathol. 45, 1893–1899 (2014).
Higgins, G. et al. Variant Ciz1 is a circulating biomarker for early-stage lung cancer. Proc. Natl. Acad. Sci. USA. 109, E3128–E3135 (2012).
Kim, S. J., Kim, E. & Rim, K. T. Non-invasive quantification of cell-free DNA mutations in plasma during lung tumor progression in mice. Cancer Biomark. 20, 477–485 (2017).
Paweletz, C. P. et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin. Cancer Res. 22, 915–922 (2016).
Ohira, T. et al. Tumor volume determines the feasibility of cell-free DNA sequencing for mutation detection in non-small cell lung cancer. Cancer Sci. 107, 1660–1666 (2016).
Yang, X. et al. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget 7, 20810–20824 (2016).
Xu, X. et al. Assessment of the clinical application of detecting EGFR, KRAS, PIK3CA and BRAF mutations in patients with non-small cell lung cancer using next-generation sequencing. Scand. J. Clin. Lab. Invest. 76, 386–392 (2016).
Jamal-Hanjani, M. et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann. Oncol. 27, 862–867 (2016).
Borsu, L. et al. Clinical Application of Picodroplet Digital PCR Technology for Rapid Detection of EGFR T790M in Next-Generation Sequencing Libraries and DNA from Limited Tumor Samples. J. Mol. Diagn. 18, 903–911 (2016).
Ma, M. et al. Comparison of plasma and tissue samples in epidermal growth factor receptor mutation by ARMS in advanced non-small cell lung cancer. Gene 591, 58–64 (2016).
Cui, S. et al. Use of capture-based next-generation sequencing to detect ALK fusion in plasma cell-free DNA of patients with non-small-cell lung cancer. Oncotarget 8, 2771–2780 (2017).
Su, D. et al. High performance of targeted next generation sequencing on variance detection in clinical tumor specimens in comparison with current conventional methods. J. Exp. Clin. Cancer Res. 36, 121 (2017).
Seki, Y. et al. Picoliter-Droplet Digital Polymerase Chain Reaction-Based Analysis of Cell-Free Plasma DNA to Assess EGFR Mutations in Lung Adenocarcinoma That Confer Resistance to Tyrosine-Kinase Inhibitors. Oncologist 21, 156–164 (2016).
Health Quality Ontario. Epidermal Growth Factor Receptor Mutation (EGFR) Testing for Prediction of Response to EGFR-Targeting Tyrosine Kinase Inhibitor (TKI) Drugs in Patients with Advanced Non-Small-Cell Lung Cancer: An Evidence-Based Analysis. Ont. Health Technol. Assess. Ser. 10, 1–48 (2010).
Won, J. K. et al. Concomitant ALK translocation and EGFR mutation in lung cancer: a comparison of direct sequencing and sensitive assays and the impact on responsiveness to tyrosine kinase inhibitor. Ann. Oncol. 26, 348–354 (2015).
Xu, S. et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett. 370, 324–331 (2016).
Drilon, A. et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin. Cancer Res. 21, 3631–3639 (2015).
Preusser, M. et al. Spectrum of gene mutations detected by next generation exome sequencing in brain metastases of lung adenocarcinoma. Eur. J. Cancer 51, 1803–1811 (2015).
Oellerich, M. et al. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit. Rev. Clin. Lab. Sci. 54, 205–218 (2017).
DiBardino, D. M. et al. Next-generation sequencing of non-small cell lung cancer using a customized, targeted sequencing panel: Emphasis on small biopsy and cytology. Cytojournal 14, doi:10.4103/1742-6413.202602 (2017).
Gao, J. et al. Comparison of Next-Generation Sequencing, Quantitative PCR, and Sanger Sequencing for Mutation Profiling of EGFR, KRAS, PIK3CA and BRAF in Clinical Lung Tumors. Clin. Lab. 62, 689–696 (2016).
Yamamoto, G. et al. Routine genetic testing of lung cancer specimens derived from surgery, bronchoscopy and fluid aspiration by next generation sequencing. Int. J. Oncol. 50, 1579–1589 (2017).
Hagemann, I. S. et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer 121, 631–639 (2015).
Qin, L., Zhong, W., Zhang, L., Li, L. Y. & Wang, M. Z. Comparison of three methods for detecting epidermal growth factor receptor mutations in plasma DNA samples of Chinese patients with advanced non-small cell lung cancer. Chin. Med. J. (Engl.) 124, 887–891 (2011).
Han, J. Y. et al. Comparison of targeted next-generation sequencing with conventional sequencing for predicting the responsiveness to epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) therapy in never-smokers with lung adenocarcinoma. Lung Cancer 85, 161–167 (2014).
Reynolds, J. P. et al. Next-generation sequencing of liquid-based cytology non-small cell lung cancer samples. Cancer 125, 178–187 (2017).
Kim, H. R. et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J. Exp. Clin. Cancer Res. 32, doi:10.1186/1756-9966-32-50 (2013).
Ishii, H. et al. Digital PCR analysis of plasma cell-free DNA for non-invasive detection of drug resistance mechanisms in EGFR mutant NSCLC: Correlation with paired tumor samples. Oncotarget 6, 30850–30858 (2015).
Youssef, O. et al. Presence of cancer-associated mutations in exhaled breath condensates of healthy individuals by next generation sequencing. Oncotarget 8, 18166–18176 (2017).
Alegre, E. et al. Total and mutated EGFR quantification in cell-free DNA from non-small cell lung cancer patients detects tumor heterogeneity and presents prognostic value. Tumour Biol. 37, 13687–13694 (2016).
Marchetti, A. et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J. Thorac. Oncol. 10, 1437–1443 (2015).
Vanni, I. et al. Next-Generation Sequencing Workflow for NSCLC Critical Samples Using a Targeted Sequencing Approach by Ion Torrent PGMTM Platform. Int. J. Mol. Sci. 16, 28765–28782 (2015).
Zhang, H. P. et al. Screening for K-ras mutations in colorectal and lung cancers by using a novel real-time PCR with ADx-K-ras kit and Sanger DNA sequencing. Zhonghua Bing Li Xue Za Zhi 39, 757–761 (2010).
Yao, Y. et al. Detection of circulating tumor DNA in patients with advanced non-small cell lung cancer. Oncotarget 8, 2130–2140 (2017).
Zhao, X. et al. Combined Targeted DNA Sequencing in Non-Small Cell Lung Cancer (NSCLC) Using UNCseq and NGScopy, and RNA Sequencing Using UNCqeR for the Detection of Genetic Aberrations in NSCLC. PLoS One 10, doi:10.1371/journal.pone.0129280 (2015).
Jiang, B. et al. Serum detection of epidermal growth factor receptor gene mutations using mutant-enriched sequencing in Chinese patients with advanced nonsmall cell lung cancer. J. Int. Med. Res. 39, 1392–1401 (2011).
Ludovini, V. et al. Plasma DNA, microsatellite alterations, and p53 tumor mutations are associated with disease-free survival in radically resected non-small cell lung cancer patients: a study of the perugia multidisciplinary team for thoracic oncology. J. Thorac. Oncol. 3, 365–373 (2008).
National Research Council (NRC). Applications of toxigenomic technologies to predictive toxicology and risk assessment. Washington, D.C.: National Academies Press (2007).
Wang, Z. et al. Global gene expression profiling in whole-blood samples from individuals exposed to metal fumes. Env. Health Perspect. 113, 233–241 (2005).
Toyoshiba, H. et al. Gene interaction network suggests dioxin induces a significant linkage between aryl hydrocarbon receptor and retinoic acid receptor beta. Environ. Health Perspect. 112, 1217–1224 (2004).
Song, M.-K., Cho, Y., Jeong, S.-C. & Ryu, J.-C. Analysis of gene expression changes in relation to hepatotoxicity induced by perfluorinated chemicals in a human hepatoma cell line. Toxicol. Environ. Health Sci. 8, 114–127 (2016).
Shostak, S. Locating gene-environment interaction: at the intersections of genetics and public health. Soc. Sci. Med. 56, 2327–2342 (2003).
Hagmar, L. et al. Impact of types of lymphocyte chromosomal aberrations on human cancer risk: results from Nordic and Italian cohorts. Cancer Res. 64, 2258–2263 (2004).
Bonassi, S., Znaor, A., Norppa, H. & Hagmar, L. Chromosomal aberrations and risk of cancer in humans: an epidemiological perspective. Cytogenet. Genome Res. 104, 376–382 (2004).
McCanlies, E. C., Kreiss, K., Andrew, M. & Weston, A. HLA-DPB1 and chronic beryllium disease: a HuGE review. Am. J. Epidemiol. 157, 388–398 (2003).
Jung, J., Hah, K., Lee, W. & Jang, W. Meta-analysis of microarray datasets for the risk assessment of coplanar polychlorinated biphenyl 77 (PCB77) on human health. Toxicol. Environ. Health Sci. 9, 161–168 (2017).
Genetics and Public Policy Center. Genetic testing practice guidelines: translating genetic discoveries into clinical care, https://doi.org/www.dnapolicy.org/resources/Genetic_Testing_Practice_Guideslines.pdf (2006).
American College of Occupational and Environmental Medicine (ACOEM). Genetic screening in the workplace, https://doi.org/www.acoem.org/guidelines.aspx?id=726 (2005).
McCanlies, E. C. et al. Impact of negatively charged patches on the surface of MHC class II antigen-presenting proteins on risk of chronic beryllium disease. J. R. Soc. Interface 5, 749–758 (2008).
Soleimani, E., Moghadam, R. H. & Ranjbar, A. Occupational exposure to chemicals and oxidative toxic stress. Toxicol. Environ. Health Sci. 7, 1–24 (2015).
Wade, P. A. & Archer, T. K. Epigenetics: environmental instructions for the genome. Environ. Health Perspect. 114, A140-A141 (2006).
Schulte, P. A. The contribution of genetics and genomics to occupational safety and health. Occup. Environ. Med. 64, 717–718 (2007).
Koizumi, S. Application of DNA microarrays in occupational health research. J. Occup. Health 46, 20–25 (2004).
Hanash S. Disease proteomics. Nature 422, 226–232 (2003).
Hunter, D. J. Gene-environment interactions in human diseases. Nat. Rev. Genet. 6, 287–298 (2005).
Barker, R. N. et al. The increase in allergic disease: environment and susceptibility. Proceedings of a symposium held at the Royal Society of Edinburgh. Clin. Exp. Allergy 33, 394–406 (2003).
74 Federal Register. 9056 [Internet]. Regulation under the genetic information nondiscrimination act of 2008, https://doi.org/edocket.access.gpo.gov/2009/E9-4221.htm (2009).
Boffetta, P. et al. Chromosomal aberrations and cancer risk: Results of a cohort study from central Europe. Am. J. Epidemiol. 165, 36–43 (2007).
Bonassi, S. et al. An increased micronucleus frequency in peripheral blood lymphocytes predicts the risk of cancer in humans. Carcinogenesis 28, 625–631 (2007).
Rossner, P. et al. Chromosomal aberration in lymphocytes of healthy subjects and risk of cancer. Env Health Persp. 113, 517–520 (2005).
Geppert, C. M. A. & Roberts, L. W. Ethical issues in the use of genetic information in the workplace: a review of recent developments. Curr. Opin. Psychiatry 18, 518–524 (2005).
Holtzman, N. A. Ethical aspects of genetic testing in the workplace. Community Genet. 6, 136–138 (2003).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rim, KT. Genetic Biomarkers and Their Applications to Prevent Occupational Diseases: A Literature Review. Toxicol. Environ. Health Sci. 10, 147–156 (2018). https://doi.org/10.1007/s13530-018-0358-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13530-018-0358-0