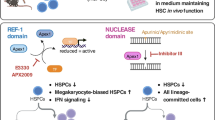
Abstract
Purpose
The eukaryotic cell plasma membrane contains several asymmetrically distributed phospholipids, which is maintained by the P4-ATPase flippase complex. Herein, we demonstrated the biological effects and mechanisms of asymmetrical loss in hematopoietic stem cells (HSCs).
Methods
An Atp8a1 knockout mouse model was employed, from which the HSC (long-term HSCs and short-term HSCs) population was analyzed to assess their abundance and function. Additionally, competitive bone marrow transplantation and 5-FU stress assays were performed. RNA sequencing was performed on Hematopoietic Stem and Progenitor Cells, and DNA damage was assayed using immunofluorescence staining and comet electrophoresis. The protein abundance for members of key signaling pathways was confirmed using western blotting.
Results
Atp8a1 deletion resulted in slight hyperleukocytosis, associated with the high proliferation of HSCs and BCR/ABL1 transformed leukemia stem cells (LSCs). Atp8a1 deletion increased the repopulation capability of HSCs with a competitive advantage in reconstitution assay. HSCs without Atp8a1 were more sensitive to 5-FU-induced apoptosis. Moreover, Atp8a1 deletion prevented HSC DNA damage and facilitated DNA repair processes. Genes involved in PI3K-AKT-mTORC1, DNA repair, and AP-1 complex signaling were enriched and elevated in HSCs with Atp8a1 deletion. Furthermore, Atp8a1 deletion caused decreased PTEN protein levels, resulting in the activation of PI3K-AKT-mTORC1 signaling, further increasing the activity of JNK/AP-1 signaling and YAP1 phosphorylation.
Conclusion
We identified the role of Atp8a1 on hematopoiesis and HSCs. Atp8a1 deletion resulted in the loss of phosphatidylserine asymmetry and intracellular signal transduction chaos.







Similar content being viewed by others
Data availability
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request. In addition, the raw sequence data reported in this paper have been deposited in the Genome Sequence Archive[28] in National Genomics Data Center[29], China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA009276), which is publicly accessible at https://ngdc.cncb.ac.cn/gsa.
This paper has not been previously published and is not under consideration by another journal. All authors have approved and agreed to submit the manuscript to this journal.
References
G. van Meer, D.R. Voelker, G.W. Feigenson, Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. (2008). https://doi.org/10.1038/nrm2330
V.V. Flis, G. Daum, Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb. Perspect. Biol. (2013). https://doi.org/10.1101/cshperspect.a013235
M. Takar, Y. Wu, T.R. Graham, The Essential Neo1 Protein from Budding Yeast Plays a Role in Establishing Aminophospholipid Asymmetry of the Plasma Membrane. J. Biol. Chem. (2016). https://doi.org/10.1074/jbc.M115.686253
N.F. Lipp, S. Ikhlef, J. Milanini, G. Drin, Lipid Exchangers: Cellular Functions and Mechanistic Links With Phosphoinositide Metabolism. Front. Cell Dev. Biol. (2020). https://doi.org/10.3389/fcell.2020.00663
T.G. Pomorski, A.K. Menon, Lipid somersaults: Uncovering the mechanisms of protein-mediated lipid flipping. Prog. Lipid Res. (2016). https://doi.org/10.1016/j.plipres.2016.08.003
D. Marquardt, B. Geier, G. Pabst, Asymmetric lipid membranes: towards more realistic model systems. Membranes (2015). https://doi.org/10.3390/membranes5020180
H.M. Hankins, R.D. Baldridge, P. Xu, T.R. Graham, Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic (2015). https://doi.org/10.1111/tra.12233
D.T. Moore, B.W. Berger, W.F. DeGrado, Protein-protein interactions in the membrane: sequence, structural, and biological motifs. Structure (2008). https://doi.org/10.1016/j.str.2008.05.007
R.L. Lopez-Marques, L. Theorin, M.G. Palmgren, T.G. Pomorski, P4-ATPases: lipid flippases in cell membranes. Pflugers Arch. (2014). https://doi.org/10.1007/s00424-013-1363-4
L.N. Bull, M.J. van Eijk, L. Pawlikowska, J.A. DeYoung, J.A. Juijn, M. Liao, L.W. Klomp, N. Lomri, R. Berger, B.F. Scharschmidt, A.S. Knisely, R.H. Houwen, N.B. Freimer, A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat. Genet. (1998). https://doi.org/10.1038/ng0398-219
M. Yabas, C.E. Teh, S. Frankenreiter, D. Lal, C.M. Roots, B. Whittle, D.T. Andrews, Y. Zhang, N.C. Teoh, J. Sprent, L.E. Tze, E.M. Kucharska, J. Kofler, G.C. Farell, S. Broer, C.C. Goodnow, A. Enders, ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat Immunol (2011). https://doi.org/10.1038/ni.2011
A.Y. Liou, L.L. Molday, J. Wang, J.P. Andersen, R.S. Molday, Identification and functional analyses of disease-associated P4-ATPase phospholipid flippase variants in red blood cells. J. Biol. Chem. (2019). https://doi.org/10.1074/jbc.RA118.007270
X. Zhu, R.T. Libby, W.N. de Vries, R.S. Smith, D.L. Wright, R.T. Bronson, K.L. Seburn, S.W. John, Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. (2012). https://doi.org/10.1371/journal.pgen.1002853
D.J. Kerr, A. Marsillo, S.R. Guariglia, T. Budylin, R. Sadek, S. Menkes, A. Chauhan, G.Y. Wen, D.P. McCloskey, A. Wieraszko, P. Banerjee, Aberrant hippocampal Atp8a1 levels are associated with altered synaptic strength, electrical activity, and autistic-like behavior. Biochem. Biophys. Acta. (2016). https://doi.org/10.1016/j.bbadis.2016.06.005
U. Kato, H. Inadome, M. Yamamoto, K. Emoto, T. Kobayashi, M. Umeda, Role for phospholipid flippase complex of ATP8A1 and CDC50A proteins in cell migration. J. Biol. Chem. (2013). https://doi.org/10.1074/jbc.M112.402701
H.W. Shin, H. Takatsu, Substrates of P4-ATPases: beyond aminophospholipids (phosphatidylserine and phosphatidylethanolamine). FASEB J. (2019). https://doi.org/10.1096/fj.201801873R
B.X. Huang, M. Akbar, K. Kevala, H.Y. Kim, Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol. (2011). https://doi.org/10.1083/jcb.201005100
H.Y. Kim, M. Akbar, A. Lau and L. Edsall, Inhibition of neuronal apoptosis by docosahexaenoic acid (22:6n-3). Role of phosphatidylserine in antiapoptotic effect. J. Biol. Chem. (2000). https://doi.org/10.1074/jbc.M004446200.
A.C. Newton, L.M. Keranen, Phosphatidyl-L-serine is necessary for protein kinase C’s high-affinity interaction with diacylglycerol-containing membranes. Biochemistry (1994). https://doi.org/10.1021/bi00187a035
K. Levano, V. Punia, M. Raghunath, P.R. Debata, G.M. Curcio, A. Mogha, S. Purkayastha, D. McCloskey, J. Fata, P. Banerjee, Atp8a1 deficiency is associated with phosphatidylserine externalization in hippocampus and delayed hippocampus-dependent learning. J. Neurochem. (2012). https://doi.org/10.1111/j.1471-4159.2011.07543.x
I. Dransfield, A. Zagorska, E.D. Lew, K. Michail, G. Lemke, Mer receptor tyrosine kinase mediates both tethering and phagocytosis of apoptotic cells. Cell Death Dis. (2015). https://doi.org/10.1038/cddis.2015.18
R.F. Zwaal, P. Comfurius, E.M. Bevers, Lipid-protein interactions in blood coagulation. Biochem. Biophys. Acta. (1998). https://doi.org/10.1016/s0304-4157(98)00018-5
P.A. Leventis, S. Grinstein, The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys (2010). https://doi.org/10.1146/annurev.biophys.093008.131234
S. Shenoy, P. Shekhar, F. Heinrich, M.C. Daou, A. Gericke, A.H. Ross, M. Losche, Membrane association of the PTEN tumor suppressor: molecular details of the protein-membrane complex from SPR binding studies and neutron reflection. PLoS One (2012). https://doi.org/10.1371/journal.pone.0032591
B.D. Manning, A. Toker, AKT/PKB Signaling: Navigating the Network. Cell (2017). https://doi.org/10.1016/j.cell.2017.04.001
F. Wu, Z. Chen, J. Liu, Y. Hou, The Akt-mTOR network at the interface of hematopoietic stem cell homeostasis. Exp. Hematol. (2021). https://doi.org/10.1016/j.exphem.2021.08.009
A. Subramanian, P. Tamayo, V.K. Mootha, S. Mukherjee, B.L. Ebert, M.A. Gillette, A. Paulovich, S.L. Pomeroy, T.R. Golub, E.S. Lander, J.P. Mesirov, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. (2005). https://doi.org/10.1073/pnas.0506580102
T. Chen, X. Chen, S. Zhang, J. Zhu, B. Tang, A. Wang, L. Dong, Z. Zhang, C. Yu and Y. Sun, The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics (2021). https://doi.org/10.1016/j.gpb.2021.08.001
Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. (2022)
J. Li, D.M. Witten, I.M. Johnstone, R. Tibshirani, Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics (2012). https://doi.org/10.1093/biostatistics/kxr031
Y. Hu, S. Li, Survival regulation of leukemia stem cells. Cell. Mol. Life Sci. (2016). https://doi.org/10.1007/s00018-015-2108-7
C. Ramkumar, R.M. Gerstein, H. Zhang, Serial transplantation of bone marrow to test self-renewal capacity of hematopoietic stem cells in vivo. Methods Mol. Biol. (2013). https://doi.org/10.1007/978-1-62703-317-6_2
D.J. Papachristou, A. Batistatou, G.P. Sykiotis, I. Varakis, A.G. Papavassiliou, Activation of the JNK-AP-1 signal transduction pathway is associated with pathogenesis and progression of human osteosarcomas. Bone (2003). https://doi.org/10.1016/s8756-3282(03)00026-7
T. Takeda, Y. Yamamoto, M. Tsubaki, T. Matsuda, A. Kimura, N. Shimo, S. Nishida, PI3K/Akt/YAP signaling promotes migration and invasion of DLD-1 colorectal cancer cells. Oncol. Lett. (2022). https://doi.org/10.3892/ol.2022.13226
M.J. Althoff, R.C. Nayak, S. Hegde, A.M. Wellendorf, B. Bohan, M.D. Filippi, M. Xin, Q.R. Lu, H. Geiger, Y. Zheng, M.T. Diaz-Meco, J. Moscat, J.A. Cancelas, Yap1-Scribble polarization is required for hematopoietic stem cell division and fate. Blood (2020). https://doi.org/10.1182/blood.2019004113
J.P. Andersen, A.L. Vestergaard, S.A. Mikkelsen, L.S. Mogensen, M. Chalat, R.S. Molday, P4-ATPases as Phospholipid Flippases-Structure, Function, and Enigmas. Front. Physiol. (2016). https://doi.org/10.3389/fphys.2016.00275
N. Li, Y. Yang, C. Liang, Q. Qiu, C. Pan, M. Li, S. Yang, L. Chen, X. Zhu, Y. Hu, Tmem30a Plays Critical Roles in Ensuring the Survival of Hematopoietic Cells and Leukemia Cells in Mice. Am. J. Pathol. (2018). https://doi.org/10.1016/j.ajpath.2018.02.015
M.P. Murphy, How mitochondria produce reactive oxygen species. Biochem. J. (2009). https://doi.org/10.1042/BJ20081386
S. Chatterjee, E.A. Browning, N. Hong, K. DeBolt, E.M. Sorokina, W. Liu, M.J. Birnbaum and A.B. Fisher, Membrane depolarization is the trigger for PI3K/Akt activation and leads to the generation of ROS. Am. J Physiol. Heart Circ Physiol (2012). https://doi.org/10.1152/ajpheart.00298.2011
Y. Shi, F. Nikulenkov, J. Zawacka-Pankau, H. Li, R. Gabdoulline, J. Xu, S. Eriksson, E. Hedstrom, N. Issaeva, A. Kel, E.S. Arner, G. Selivanova, ROS-dependent activation of JNK converts p53 into an efficient inhibitor of oncogenes leading to robust apoptosis. Cell Death Differ. (2014). https://doi.org/10.1038/cdd.2013.186
E.H. Blackburn, C.W. Greider, J.W. Szostak, Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. (2006). https://doi.org/10.1038/nm1006-1133
T. Matsudaira, K. Mukai, T. Noguchi, J. Hasegawa, T. Hatta, S.I. Iemura, T. Natsume, N. Miyamura, H. Nishina, J. Nakayama, K. Semba, T. Tomita, S. Murata, H. Arai, T. Taguchi, Endosomal phosphatidylserine is critical for the YAP signalling pathway in proliferating cells. Nat Commun. (2017). https://doi.org/10.1038/s41467-017-01255-3
T. Konishi, R.M. Schuster and A.B. Lentsch, Proliferation of hepatic stellate cells, mediated by YAP and TAZ, contributes to liver repair and regeneration after liver ischemia-reperfusion injury. American journal of physiology. Gastrointest. Liver Physiol. (2018). https://doi.org/10.1152/ajpgi.00153.2017
C. Dai, X. Chen, J. Li, P. Comish, R. Kang, D. Tang, Transcription factors in ferroptotic cell death. Cancer Gene Ther. (2020). https://doi.org/10.1038/s41417-020-0170-2
J. Rosenbluh, D. Nijhawan, A.G. Cox, X. Li, J.T. Neal, E.J. Schafer, T.I. Zack, X. Wang, A. Tsherniak, A.C. Schinzel, D.D. Shao, S.E. Schumacher, B.A. Weir, F. Vazquez, G.S. Cowley, D.E. Root, J.P. Mesirov, R. Beroukhim, C.J. Kuo, W. Goessling, W.C. Hahn, beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell (2012). https://doi.org/10.1016/j.cell.2012.11.026
O.P. Pinto do, K. Richter and L. Carlsson, Hematopoietic progenitor/stem cells immortalized by Lhx2 generate functional hematopoietic cells in vivo. Blood (2002). https://doi.org/10.1182/blood.v99.11.3939
K. Richter, V. Wirta, L. Dahl, S. Bruce, J. Lundeberg, L. Carlsson, C. Williams, Global gene expression analyses of hematopoietic stem cell-like cell lines with inducible Lhx2 expression. BMC Genomics (2006). https://doi.org/10.1186/1471-2164-7-75
Acknowledgements
The authors greatly appreciate the State Key Laboratory of Biotherapy & Collaborative Innovation Center for Biotherapy for their support, the staff of the core facility and the animal facility of the State Key Laboratory of Biotherapy and West China Hospital.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82170114 to Y. Hu) and National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University (Z20201008 to Y. Hu), Guizhou Provincial Science & Technology Support Program (NO [2020]4Y061 to Y. Hu). National Natural Science Foundation of China (81802468 to LZh). Chengdu Science and Technology innovation project (2021-YF05-00800-SN to LZh).
Author information
Authors and Affiliations
Contributions
L.Z, C.P and Y.H conceived research ideas, designed experiments, analyzed data, and wrote the manuscript. L.Z., W.T. and C.L. performed experiments; Y.F and B.W performed the bioinformatics; H. Q, Q.Q, N.L; W.H. Y.S and Z. Y helped complete the experiments. L.Z, X.Z and K.S reviewed and edited the manuscript, discussed the results and commented on the manuscript. All authors have read and approved the article.
Corresponding authors
Ethics declarations
Ethical approval
All mouse studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the Sichuan University. All animals were monitored for abnormal behavior to minimize pain and suffering. Animals were euthanized when excessive deterioration of health was noted.
Competing interests
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, L., Pan, C., Tian, W. et al. Atp8a1 deletion increases the proliferative activity of hematopoietic stem cells by impairing PTEN function. Cell Oncol. 46, 1069–1083 (2023). https://doi.org/10.1007/s13402-023-00797-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-023-00797-7