Abstract
The small Indian mongoose, Urva auropunctata, is one of the most notorious invasive species in the world. In Japan, the mongoose was introduced to Amami, Kyushu (Kiire region), and Okinawa Islands. Currently, an eradication program for the Amami population is proving successful. To prevent reinvasion and conserve biodiversity, it would be advantageous to investigate the origin and dispersal history of introduced mongoose populations in Japan. We determined the mitochondrial genome of a mongoose from the Okinawa population and confirmed that the systematic status of the mongoose is Urva auropunctata. We also found two haplotypes of cytochrome b in the Amami population, one of which is common among the Okinawa, Amami, and Kiire populations. Based on our microsatellite analysis, the three populations were genetically different from each other. Furthermore, with a low pairwise FST value, the Kiire population was closer to the Okinawa than the Amami population. Our results suggest that the Okinawa population could be the source of the Amami and Kiire populations. These results align with historical records of mongoose in Japan and clarify its genetic status. Our results should aid the prediction of the pathway through which a new invasion will likely occur.
Similar content being viewed by others
Introduction
Invasive non-native species are among the greatest threats to biodiversity around the world (Bellard et al. 2016). In particular, invasive species can easily establish populations on islands, as their endemic ecosystems were established over a long evolutionary history and more niche space is available compared with complex ecosystems (Bellard et al. 2014; Kappes et al. 2021). Revealing the invasion pathways of non-native species is essential for preventing biological invasions (Hulme 2009). Recently, genetic analysis has become vital to the management of invasive species, as it provides information not only about the genetic diversity of an invasive species, but also its origin and introduction history (Croucher et al. 2013; Browett et al. 2020). Such analyses are effective for identifying eradication and control units (Robertson and Gemmell 2004; Abdelkrim et al. 2005; Fraser et al. 2013).
The small Indian mongoose Urva auropunctata (Hodgson 1836) is a generalist predator with a native range from the Arabian Peninsula to Southeast Asia (Gilchrist et al. 2009). The systematic status of this mongoose has been confused; it was known as Herpestes javanicus and H. auropunctatus before its systematic status and biogeography were genetically clarified using partial sequences of mitochondrial DNA (Veron et al. 2007; Patou et al. 2009; Veron and Jennings 2017). The mongoose was introduced to 64 islands (Pacific and Indian Oceans, Caribbean, and Adriatic Seas) and continental areas (Europe, South America, Australia, and North America) through human activities for rat and snake control, and invasive mongooses severely impacted native biodiversity (Hays and Conant 2007; Watari et al. 2008; Barun et al. 2011). The introduction histories of this species have been thoroughly investigated based on genetic inference and historical records around the world (e.g., Thulin et al. 2006; Veron et al. 2007; Barun et al. 2013; Wostenberg et al. 2019; Louppe et al. 2021). In addition, ecological niche modeling for the mongoose suggests that the distribution of invasive mongooses will expand substantially by 2050 (Louppe et al. 2020).
In Japan, the mongoose was introduced for rat and snake control and has become a serious threat to biodiversity. Initially, 13–17 mongooses were introduced to Okinawa Island (Okinawa) and 4 mongooses were introduced to Tonaki Island from Bangladesh in 1910 (Kishida 1931; Yamada et al. 2015). Around 1954, approximately 30 individuals were introduced to Ie-jima Island and 9 individuals were introduced to Ishigaki Island from Okinawa Island (Iha 1966). On Amami-Ohshima Island (Amami), approximately 30 individuals were introduced around 1979 (Abe et al. 1991; Yamada et al. 2015). Since then, the mongooses have expanded their distribution range, except on Tonaki, Ie-jima, and Ishigaki islands, where the populations failed to colonize (Kishida 1931; Iha 1966; Ishibashi and Ogura 2012). Recently, colonization by the mongoose was reported in the Kiire region of Kyushu Island (Kiire), which is one of the main Japanese islands, where it has been established for at least 30 years (Watari et al. 2011). A mongoose eradication program is currently ongoing in Japan, which is proving successful and has improved conditions for native species in Amami (Fukasawa et al. 2013b; Watari et al. 2013; Watari 2019). The Kiire population has also virtually disappeared, based on trapping and auto-sensor camera surveys (Funakoshi et al. 2015). To prevent reinvasion and conserve biodiversity, it would be worthwhile to investigate the origins and dispersal history of these introduced populations in Japan.
Genetic analyses of mongooses in Japan have been reported previously. The same haplotypes of partial sequences of mitochondrial DNA (mtDNA) cytochrome b (Cytb) were found in the Okinawa, Amami, and Kiire populations (Sekiguchi et al. 2001; Watari et al. 2011). Although the analysis of the mitochondrial genome (mitogenome) provides more accurate phylogenetic results than the analysis of partial mtDNA with decreased stochastic errors (Li et al. 2014), the mitogenome of Japanese mongoose has not yet been determined. In addition, using clustering analysis based on eight microsatellite loci, Thulin et al. (2006) showed that the Okinawa and Amami populations have a recent common history. However, the origin of the Kiire population has not yet been investigated, and three of Thulin’s eight microsatellite loci could not be scored reliably, making it difficult to reject alternative hypotheses in other regions (Barun et al. 2013). Thus, the origin and genetic diversity of mongoose populations in Japan require reevaluation and discussion.
In the present study, we investigated the phylogenetic status of a mongoose in Japan based on its mitogenome sequence and analyzed the population structure of Japanese mongooses using the complete sequences of Cytb and 17 microsatellite loci. These data are used to discuss the genetic characters and origins of mongoose populations in Japan.
Materials and methods
Samples and DNA extraction
We collected 74 mongoose tissue samples (Table S1, Fig. 1), including 28 from Okinawa, 40 from Amami, and 6 from Kiire. These samples were obtained from mongoose eradication programs conducted on Amami and Okinawa islands and in collaborative laboratories. The 28 samples from Okinawa were also analyzed by Sato and Jogahara (2021). Total DNA was extracted from the samples using the DNeasy Tissue and Blood Kit (QIAGEN, Hilden, Germany). For samples of unknown sex, we used the sex determination method of Sato and Jogahara (2021).
Analysis of the mitogenome of the small Indian mongoose
One mitogenome was assembled from whole-genome sequencing data (DRR294934) obtained from a mongoose (Nago1) of the Okinawa population (Sato and Jogahara 2021). The low-quality bases and adapters of the sequence were trimmed using fastp (Chen et al. 2018) with the default configuration. The mitogenome was assembled using GetOrganelle v1.7.5 (Jin et al. 2020) and annotated using MITOS (Bernt et al. 2013). We compared the mitogenome with 14 other sequences of Herpestidae obtained from Hassanin et al. (2021), and overlapping regions were used for subsequent analysis. The final alignment contains 15 sequences and has a length of 14,851 bp. To determine the phylogenetic status of the invasive mongoose in Japan, we constructed a maximum likelihood (ML) tree based on the alignment obtained with the GTR (General Time Reversible) model (Tavaré, 1986) with a gamma distribution (+ G) and invariant sites (+ I) using MEGA7 (Kumar et al. 2016). The phylogeny was tested with 1000 bootstrap replicates.
Analysis of cytochrome b
To amplify the complete sequence of Cytb (1140 bp), two new primers were designed: moncb_F1, 5ʹ-CACATGGAATCTAACCATGAC-3ʹ, and moncb_R1, 5ʹ-CTTGAAGTCTTAGGGAGG-3ʹ. Polymerase chain reaction (PCR) was conducted in a volume of 10 μl including 2.0 μl of 5 × Prime STAR GXL DNA Buffer (Takara, Shiga, Japan), 0.8 μl of dNTP mixture (2.5 mM each dNTP; Takara), 0.2 μl of Prime STAR GXL DNA Polymerase (1.25 U/μl; Takara), 0.4 μl each of the forward and reverse primers (10 pmol/μl), and 1.0 μl of the DNA extract. The volume was adjusted to 10 μl with distilled water. Amplification was performed at 98 °C for 1 min, followed by 35 cycles of 98 °C for 10 s, 56.5 °C for 15 s, and 68 °C for 1 min using a MiniAmp Plus thermal cycler (Applied Biosystems [ABI], Waltham, MA, USA). The amplicons were checked through electrophoresis on 2% agarose gels. We also confirmed the lack of PCR amplification in the negative controls. The PCR products were purified using ExoSAP-IT Express (ABI) reagent and sequenced by Macrogen (Kyoto, Japan).
We compared our Cytb sequences with those of H. javanicus and U. auropunctata obtained from a public database, as the small Indian mongoose was formerly designated H. javanicus (Patou et al. 2009). Here, we used only complete Cytb sequences with no ambiguous sites. The final alignment contains 127 sequences and 1140 bp; 53 sequences were obtained from Ledje and Arnason (1996), Yoder et al. (2003), Koepfli et al. (2006), Veron et al. (2007), Louppe et al. (2021), Hassanin et al. (2021), and Penny and McLenachan (unpublished; Table S2). The haplotypes were detected using the TCS program (Clement et al. 2000) and a median-joining network was constructed with PopART (Leigh and Bryant 2015).
Genotyping of microsatellite loci
According to the method of Sato and Jogahara (2021), we genotyped 17 microsatellite loci (Uau1–17) with six multiplex sets (s1: Uau1, 11, 17; s2: Uau2, 7, 13; s3: Uau3, 9, 14; s4: Uau4, 10, 15; s5: Uau8, 16; s6: Uau5, 6, 12). Fragment analysis was conducted by the FASMAC DNA sequencing service (http://fasmac.co.jp/) on an ABI 3730xl DNA Analyzer with GeneScan 600 LIZ Size Standard (ABI). For genotyping, Peakscanner Software 1.0 (ABI) was used; we assessed the repeatability of heterozygous genotypes twice and homozygous genotypes three times. Data for the Okinawa population were obtained from Sato and Jogahara (2021).
Genetic diversity and population differentiation
For all microsatellite loci in each population, scoring error, allele dropout, and the presence of null alleles were tested using MICRO-CHECKER version 2.2 (Van Oosterhout et al. 2004). The deviation from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) of each locus were determined using GENEPOP v4.2 (Raymond and Rousset 1995). The number of alleles (k), observed heterozygosity (HO), expected heterozygosity (HE), pairwise FST, and absolute number of migrants (M) between populations were calculated using ARLEQUIN ver. 3.5.1.2 (Excoffier and Lischer 2010). In addition, private alleles (PA) were searched for using the poppr package (Kamvar et al. 2014) for R (R Core Team 2021), and the deviation from HWE and inbreeding coefficient (FIS) of each population were examined using GENEPOP and ARLEQUIN, respectively.
To investigate the genetic structure of the populations, we employed three approaches based on microsatellites. Principal coordinate analysis (PCoA) was conducted using GENALEX (Peakall and Smouse 2006), and discriminant analysis of principal components (DAPC) was performed using the adegenet package (Jombart 2008) in R. PCoA focuses on the total variance, and DAPC maximizes differences among populations while minimizing variation within groups. Lastly, we applied a Bayesian clustering method using STRUCTURE v2.3.4 (Pritchard et al. 2000) with a burn-in phase of 5 × 105 iterations followed by a run phase of 105 iterations. An admixture model with 5 repetitions per simulation was used to determine the number of genetic clusters (K; from 1 to 10). The optimal K was determined using the log-likelihood ratio [L (K)] and delta K were obtained using STRUCTURE HARVESTER v.0.6.94 (Earl and vonHoldt 2012).
History of Japanese mongoose populations
To confirm the introduction history of each population, population bottlenecks in all three populations were estimated using BOTTLENECK v.1.2.02 (Piry et al. 1999) with three models, i.e., an infinite allele model (IAM), two-phase mutation model (TPM), and stepwise mutation model (SMM), based on microsatellite data. For TPM, we set the variance to 12 and the proportion of SMM to 78%.
To calculate contemporary migration rates among the three populations, we used BayesAss 3.0 (Wilson and Rannala 2003). BayesAss provides migration rates over the last few generations. Estimation was performed with the following configuration: 107 iterations with a burn-in period of 106 iterations and a sampling frequency of 1000. The mixing parameters DeltaA, DeltaF, and DeltaM were set to 0.30, 0.50, and 0.40, respectively. Convergence was validated for each parameter using Tracer v1.7 (Rambaut et al. 2018). Three runs were performed to confirm the consistency of the results among runs.
Results
Phylogenetic position of Japanese mongooses based on the mitogenome
The mitogenome of a mongoose from Okinawa with a length of 16,733 bp was successfully produced, and the sequence was deposited into the DNA Data Bank of Japan (DDBJ) under accession number LC642815. The ML tree shows that our mitogenome lineage clusters with the Fijian mongoose lineage (accession number NC_006835), indicating that the mongoose in Japan is Urva auropunctata (Fig. 2).
Maximum likelihood (ML) tree of the mitogenomes of Herpestidae. The ML tree was based on the GTR + G + I model. The bootstrap support values are shown at nodes. Our sequence is shown in bold (LC642815), and the other data from Hassanin et al. (2021) are shown with accession numbers
Genetic diversity based on Cytb and microsatellites
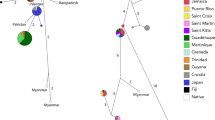
We determined 73 sequences of Cytb (1140 bp) and deposited them into DDBJ under accession numbers LC684938 to LC685010. Based on TCS analysis, two Cytb haplotypes are present in Japan: UaC6 and UaC10 (Table S1, Fig. 3). UaC6 was distributed among all three populations, while UaC10 was observed only in the Amami population. These two haplotypes differ by one nucleotide substitution.
Haplotype network of mtDNA Cytb. Asterisks show the haplotypes observed in our samples. The size of each circle indicates the proportion of haplotype frequencies. Black circles are estimated haplotypes, and a hatch mark between haplotypes represents a nucleotide substitution. The dashed line divides U. auropunctata (left) from U. javanica (right). The data obtained from previous studies are listed in Table S2. The Cytb haplotype of our mitogenome sequence from Okinawa (LC642815) is UaC6, Fiji’s mongoose mitogenome (NC_006835) is UaC8, and Thailand’s mitogenome (MW257214) is UjC6
All 17 microsatellite loci were successfully genotyped in all mongoose individuals (Supplemental file1), and MICRO-CHECKER did not indicate the scoring error, the allele dropout, or the presence of null alleles in our data. Three loci (Uau1, 15, and 16) deviated from HWE and were removed from subsequent analyses (Table S3). Based on the LD tests, 19 pairs of loci were likely linked (Table S4), and the linkage was commonly observed in the Amami population. This finding suggests that the effects of evolutionary forces other than linkage, due to genomic proximity between loci, are at play in each population. Therefore, these loci were included in subsequent analyses, and all further analyses were based on 14 loci (Supplemental file2).
The genetic diversity indices of mongoose populations in Japan are shown in Table 1. The allele number (k) was 2.00–2.71. One PA was found in each of the Okinawa and Amami populations. The HO and HE values were 0.42–0.54 and 0.40–0.54, respectively. The Okinawa population had the highest heterozygosity (HO, 0.54). All populations were in HWE and showed no inbreeding, with low FIS values (− 0.117 to 0.049).
Population differentiation
Our PCoA result indicates that the Okinawa and Amami populations are clearly differentiated, and the Kiire population is relatively close to the Okinawa population; however, the amount of variance explained among all populations was small (the top two principal components explained only 17.34%) (Fig. 4a). In the DAPC result, all populations were clearly distinguished (Fig. 4b). In the STRUCTURE analysis, K = 2 was optimal; the L (K) and delta K values are shown in Fig. S1. At K = 2 and 3, the Okinawa and Amami populations were distinguished and the Kiire population is genetically close to the Okinawa population (Fig. 5). In both cases, three individuals of the Amami population showed genetic characters similar to the Okinawa population. Based on pairwise FST values, the three populations differed genetically (Table 2), and the Kiire population was closer to the Okinawa (FST = 0.115) than the Amami (FST = 0.156) population.
Population history
BOTTLENECK analysis of the introduction history indicated that the number of founders for all three populations was small (Table S5). A bottleneck of the Okinawa population was detected in all models, and a bottleneck for Amami was also detected with IAM and TPM. Although the sample comprised only six individuals, a bottleneck for Kiire was supported by IAM analysis.
No contemporary migration histories were detected among the three populations by BayesAss analysis (Table S6).
Discussion
Phylogenetic status and mtDNA variations
The systematic status of the Japanese mongoose has been a source of confusion (Abe et al. 1991; Ogura et al. 1998; Nagayama et al. 2001; Sekiguchi et al. 2001), as with other mongoose populations around the world (Veron et al. 2007; Patou et al. 2009; Veron and Jennings 2017). Based on our mitogenome and Cytb results (Figs. 2 and 3), the Japanese mongoose was confirmed to be the small Indian mongoose, in accordance with Sekiguchi et al. (2001) and Watari et al. (2011). Moreover, two Cytb haplotypes are specific to Japan (Fig. 3). One of these haplotypes (UaC6) is distributed across all three populations (Okinawa, Amami, and Kiire), while the other one (UaC10) was observed only in the Amami population. UaC10 could be derived from UaC6, or may not have been observed in other populations due to chance; the difference between these two haplotypes is only one substitution. These mitochondrial analyses show that the mongooses in Japan could have originated from small founder populations. This is supported by the documented introduction history of the mongoose in Japan: mongooses in Japan originated from 13–17 individuals introduced to Okinawa Island (Kishida 1931), and the founders of the Amami population were from the Okinawa population (Abe et al. 1991; Yamada et al. 2015).
Genetic relationship between Okinawa and Amami populations
The steppingstone introduction of mongooses from Bangladesh to Okinawa, and then to Amami, was ascertained by Thulin et al. (2006) and Barun et al. (2013). The steppingstone introduction between Okinawa and Amami was also supported by our results. The allele numbers and HO values of samples from Okinawa were higher than those of the Amami and Kiire populations (Table 1). The pairwise FST value was lowest between the Okinawa and Amami populations, and the number of migrants (M) was highest between the same populations (Table 2). Thus, our genetic results agree with previous reports that Okinawa is the source of the mongoose populations in Japan and that Amami mongooses originated from the Okinawa population (Kishida 1931; Abe et al. 1991; Yamada et al. 2015).
Island populations are easily affected by genetic drift and founder effects (Clegg et al. 2002). The Okinawa and Amami populations have a bottleneck history with no recent migration between populations (Tables S5 and S6). Thus, these populations are typical island populations that originated from small numbers of founders and are isolated from each other. Thulin et al. (2006) and Louppe et al. (2021) demonstrated that the dispersal ability of the mongoose is high within islands and limited between islands. Considering the population history, this limitation is sufficient to cause a genetic difference between populations, as the investigated populations were established around 43–112 years ago. The year of introduction was 1910 for Okinawa, around 1979 for Amami, and around 1979 for the Kiire population (Kishida 1931; Watari et al. 2011; Yamada et al. 2015).
As expected, multiple analyses showed that the Okinawa and Amami populations are genetically distinct (Figs. 4 and 5, Table 2). However, Thulin et al. (2006) showed that Okinawa and Amami populations were genetically similar, with a low pairwise FST value (0.059), while Thulin’s clustering analysis showed that the Okinawa and Amami populations were well-mixed. Our FST value was 0.107 and clustering analysis distinguished the Okinawa and Amami populations (Figs. 4 and 5, Table 2). The genetic difference may have been greater in our analysis than that of Thulin et al. (2006) because the number of analyzed microsatellite loci was increased from 8 to 14, allowing us to obtain more detailed information. The FIS value for Okinawa in our study (0.011) was lower than that (0.102) reported by Thulin et al. (2006), despite the number of analyzed samples decreasing from 94 to 28. Other plausible reasons are population reductions and sampling biases. On Amami-Ohshima Island, a large-scale and long-term mongoose eradication program is ongoing and is currently proving successful (Watari 2019). The population size was estimated as 6141 in 2000 (Fukasawa et al. 2013a), and the rapid reduction thereof may impact the Amami population. This possibility is supported by our analysis of LD, which was commonly observed in the Amami population (Table S4). Location-dependent linkage is often discussed as an effect of some evolutionary forces (Urquía et al. 2019), such as genetic drift and founder effect. We may have been unable to reveal all genetic characteristics of the Okinawa population because our sampling was biased toward the northern part of Okinawa, far from the area where the mongoose was introduced. This possibility should be investigated through more comprehensive sampling in Okinawa.
Origin of the Kiire population
Watari et al. (2011) suggested that the Kiire population could have originated from released or fugitive individuals from facilities where live shows pitting mongooses against venomous snakes were previously held around the Kiire area. This suggests that the founder population of Kiire was small. A small founder population is supported by our genetic analyses: Kiire had the lowest genetic diversity and its population had a bottleneck history (Tables S5 and S6). Moreover, the Kiire population is genetically closer to the Okinawa population (Figs. 4 and 5, Table 2). According to a showman who performed such live shows in Amami-Ohshima, another showman brought mongooses from Okinawa Prefecture to Kagoshima Prefecture (Asahi Shinbun Newspaper, 2010). This account, together with and our results, suggests that the Kiire population may have originated from the Okinawa population. To validate this, more samples from the Kiire region are needed, including from other parts of Kagoshima Prefecture. However, it is difficult to obtain more mongoose samples because there have been no records of mongoose in Kagoshima since 2016, when their presence was last recorded (Nishi Nippon Shinbun Newspaper 2016).
Conclusion
The first mitogenome of the small Indian mongoose from Japan was obtained in the present study, and its phylogenetic status was verified through reconstruction of the phylogenetic tree, which confirmed that the mongoose in Japan is the small Indian mongoose, U. auropunctata. Our mtDNA and microsatellite analyses revealed the genetic relationships among the Okinawa, Amami, and Kiire populations in Japan in detail. This is the first investigation of the genetic diversity of the Kiire population performed using microsatellites. We demonstrated genetic separation between the Okinawa and Amami populations. As a result, we verified the introduction history of the small Indian mongoose from Okinawa to Amami and proposed that the Kiire population originated from Okinawa.
Revealing the introduction pathway of an invasive species is critical for the evaluation and management of new invasion risks. At present, the mongoose eradication program in Okinawa is conducted only in the northern part of Okinawa Island. No biosecurity measures have been implemented against the mongoose in port areas where vessels connect Okinawa Island to Amami-Oshima and Kyushu Islands, although no reports of mongoose detection at the ports are available. Our results did not indicate gene flow driven by migration between the islands after the introduction events. However, a new invasion of mongooses was confirmed between the Hawaiian Islands in 2012 (Wostenberg et al. 2019). Thus, a new invasion could also occur in Japan. When the new invasion occurs, examining the genetic characteristics of the invaders using our genetic markers and comparing our results may help identify the origin and pathway of the invaders. It will contribute to the biosecurity measure of mongooses.
Data availability
DNA sequences are deposited in DDBJ (LC642815 and LC684938–LC685010). Microsatellite data are available in Supplementary files 1 and 2.
References
Abdelkrim J, Pascal M, Calmet C, Samadi S (2005) Importance of assessing population genetic structure before eradication of invasive species: examples from insular Norway rat populations. Conserv Biol 19:1509–1518. https://doi.org/10.1111/j.1523-1739.2005.00206.x
Abe S, Takatsuki Y, Handa Y, Nigi H (1991) Establishment in the wild of the mongoose (Herpestes sp.) on Amami-oshima Island. Honyurui Kagaku (Mammalian Science) 31:23–36. https://doi.org/10.11238/mammalianscience.31.23 (in Japanese with English summary)
Barun A, Hanson CC, Campbell KJ, Simberloff D (2011) A review of small Indian mongoose management and eradications on islands. In: Veitch CR, Clout MN, Towns DR (eds) Island invasives: eradication and management. IUCN (International Union for Conservation of Nature), Gland, pp 17–25
Barun A, Niemiller ML, Fitzpatrick BM et al (2013) Can genetic data confirm or refute historical records? The island invasion of the small Indian mongoose (Herpestes auropunctatus). Biol Invasions 15:2243–2251. https://doi.org/10.1007/s10530-013-0447-6
Bellard C, Leclerc C, Leroy B et al (2014) Vulnerability of biodiversity hotspots to global change. Glob Ecol Biogeogr 23:1376–1386. https://doi.org/10.1111/geb.12228
Bellard C, Cassey P, Blackburn TM (2016) Alien species as a driver of recent extinctions. Biol Lett 12:20150623. https://doi.org/10.1098/rsbl.2015.0623
Bernt M, Donath A, Jühling F et al (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol 69:313–319. https://doi.org/10.1016/j.ympev.2012.08.023
Browett SS, O’Meara DB, McDevitt AD (2020) Genetic tools in the management of invasive mammals: recent trends and future perspectives. Mamm Rev 50:200–210. https://doi.org/10.1111/mam.12189
Chen S, Zhou Y, Chen Y, Gu J (2018) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Clegg SM, Degnan SM, Kikkawa J et al (2002) Genetic consequences of sequential founder events by an island-colonizing bird. Proc Natl Acad Sci 99:8127–8132. https://doi.org/10.1073/pnas.102583399
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659. https://doi.org/10.1046/j.1365-294X.2000.01020.x
Croucher PJP, Mascheretti S, Garbelotto M (2013) Combining field epidemiological information and genetic data to comprehensively reconstruct the invasion history and the microevolution of the sudden oak death agent Phytophthora ramorum (Stramenopila: Oomycetes) in California. Biol Invasions 15:2281–2297. https://doi.org/10.1007/s10530-013-0453-8
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Fraser EJ, Macdonald DW, Oliver MK et al (2013) Using population genetic structure of an invasive mammal to target control efforts - an example of the American mink in Scotland. Biol Conserv 167:35–42. https://doi.org/10.1016/j.biocon.2013.07.011
Fukasawa K, Hashimoto T, Tatara M, Abe S (2013a) Reconstruction and prediction of invasive mongoose population dynamics from history of introduction and management: a Bayesian state-space modelling approach. J Appl Ecol 50:469–478. https://doi.org/10.1111/1365-2664.12058
Fukasawa K, Miyashita T, Hashimoto T et al (2013b) Differential population responses of native and alien rodents to an invasive predator, habitat alteration and plant masting. Proc R Soc B Biol Sci 280:20132075. https://doi.org/10.1098/rspb.2013.2075
Funakoshi K, Okada S, Nagasato A, Arai A (2015) Probable achievement of eradication of the small Indian mongoose Herpestes auropunctatus and survival state of native animals in Kagoshima prefecture mainland, Japan. Honyurui Kagaku (Mammalian Science) 55:167–181. https://doi.org/10.11238/mammalianscience.55.167. (in Japanese with English summary)
Gilchrist JS, Jennings AP, Veron G, Cavallini P (2009) Family Herpestidae (Mongooses). In: Wilson DE, Mittermeier RA (eds) Handbook of the mammals of the world: Vol. 1, Carnivores. pp 222–329
Hassanin A, Veron G, Ropiquet A et al (2021) Evolutionary history of Carnivora (Mammalia, Laurasiatheria) inferred from mitochondrial genomes. Plos One 16:e0240770. https://doi.org/10.1371/journal.pone.0240770
Hays WST, Conant S (2007) Biology and impacts of Pacific Island invasive species. 1. A worldwide review of effects of the small Indian mongoose, Herpestes javanicus (Carnivora: Herpestidae). Pacific Sci 61:3–16. https://doi.org/10.1353/psc.2007.0006
Hulme PE (2009) Trade, transport and trouble: managing invasive species pathways in an era of globalization. J Appl Ecol 46:10–18. https://doi.org/10.1111/j.1365-2664.2008.01600.x
Iha K (1966) Distribution and food habits of mongoose in Okinawa. J Okinawa Agric 5:39–44 (in Japanese)
Ishibashi O, Ogura G (2012) Present situation of mongoose in Japan and survey of Leptospira in mongoose. Chikyu Kankyo 17:193–202 (in Japanese)
Jin JJ, Bin YuW, Yang JB et al (2020) GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol 21:1–31. https://doi.org/10.1186/s13059-020-02154-5
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/BIOINFORMATICS/BTN129
Kamvar ZN, Tabima JF, Grunwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 24:1–14. https://doi.org/10.7717/peerj.281
Kappes PJ, Benkwitt CE, Spatz DR, et al. (2021) Do invasive mammal eradications from islands support climate change adaptation and mitigation? Climate 9:. https://doi.org/10.3390/cli9120172
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Kishida K (1931) Professor Watase and import of mongoose. Zool Mag 43:70–78 (in Japanese)
Koepfli KP, Jenks SM, Eizirik E et al (2006) Molecular systematics of the Hyaenidae: relationships of a relictual lineage resolved by a molecular supermatrix. Mol Phylogenet Evol 38:603–620. https://doi.org/10.1016/j.ympev.2005.10.017
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Ledje C, Arnason U (1996) Phylogenetic analyses of complete cytochrome b genes of the order carnivora with particular emphasis on the Caniformia. J Mol Evol 42:135–144. https://doi.org/10.1007/BF02198839
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116. https://doi.org/10.1111/2041-210X.12410
Li B, Wolsan M, Wu D et al (2014) Mitochondrial genomes reveal the pattern and timing of marten (Martes), wolverine (Gulo), and fisher (Pekania) diversification. Mol Phylogenet Evol 80:156–164. https://doi.org/10.1016/j.ympev.2014.08.002
Louppe V, Leroy B, Herrel A, Veron G (2020) The globally invasive small Indian mongoose Urva auropunctata is likely to spread with climate change. Sci Rep 10:1–11. https://doi.org/10.1038/s41598-020-64502-6
Louppe V, Lalis A, Abdelkrim J et al (2021) Dispersal history of a globally introduced carnivore, the small Indian mongoose Urva auropunctata, with an emphasis on the Caribbean region. Biol Invasions 23:2573–2590. https://doi.org/10.1007/s10530-021-02523-6
Nagayama Y, Ogura G, Kawashima Y (2001) Morphometry of skulls and statistical verification of mongoose (Herpestes javanicus) on Okinawa and Amami Ohshima Islands. Honyurui Kagaku (Mammalian Science) 41:159–169. https://doi.org/10.11238/mammalianscience.41.159. (in Japanese with English summary)
Ogura G, Sakashita M, Kawashima Y (1998) External morphology and classification of mongoose on Okinawa Island. Honyurui Kagaku (Mammalian Science) 38:259–270. https://doi.org/10.11238/mammalianscience.38.259. (in Japanese with English summary)
Patou ML, McLenachan PA, Morley CG et al (2009) Molecular phylogeny of the Herpestidae (Mammalia, Carnivora) with a special emphasis on the Asian Herpestes. Mol Phylogenet Evol 53:69–80. https://doi.org/10.1016/j.ympev.2009.05.038
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503. https://doi.org/10.1093/jhered/90.4.502
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
Rambaut A, Drummond AJ, Xie D et al (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. https://doi.org/10.1093/SYSBIO/SYY032
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249. https://doi.org/10.1093/oxfordjournals.jhered.a111573
Robertson BC, Gemmell NJ (2004) Defining eradication units to control invasive pests. J Appl Ecol 41:1042–1048. https://doi.org/10.1111/j.0021-8901.2004.00984.x
Sato T, Jogahara T (2021) Development and characterization of microsatellite markers in the small Indian mongoose (Urva auropunctata). Mol Biol Rep 48:7029–7034. https://doi.org/10.1007/s11033-021-06655-9
Sekiguchi K, Inoue F, Ueda T et al (2001) Genealogical relationship between introduced mongooses in Okinawa and Amamiohsima Islands, Ryukyu Archipelago, inferred from sequences of mtDNA cytochrome b gene. Honyurui Kagaku (Mammalian Science) 41:65–70. https://doi.org/10.11238/mammalianscience.41.65 (in Japanese with English summary)
Tavaré S (1986) Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci 17:57–86
The Asahi Shinbun Newspaper. 2010. The cold temperature increased the body weight of the mongoose in Kagoshima prefecture: changes in body shape. 6 March 2010. https://www.asahi.com/eco/SEB201003060004.html
The Nishi Nippon Shinbun Newspaper. 2016. It should be “eradication of the mainland”…road kills and feces. Mongoose invisible. 3 months capture operation without clues. At Minami-Satsuma city, Kagoshima. 24 August 2016. https://www.nishinippon.co.jp/item/o/276412/
Thulin CG, Simberloff D, Barun A et al (2006) Genetic divergence in the small Indian mongoose (Herpestes auropunctatus), a widely distributed invasive species. Mol Ecol 15:3947–3956. https://doi.org/10.1111/j.1365-294X.2006.03084.x
Urquía D, Gutierrez B, Pozo G et al (2019) Psidium guajava in the Galapagos Islands: population genetics and history of an invasive species. Plos One 14:e0203737. https://doi.org/10.1371/journal.pone.0203737
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Veron G, Jennings AP (2017) Javan mongoose or small Indian mongoose–who is where? Mamm Biol 87:62–70. https://doi.org/10.1016/j.mambio.2017.05.006
Veron G, Patou ML, Pothet G et al (2007) Systematic status and biogeography of the Javan and small Indian mongooses (Herpestidae, Carnivora). Zool Scr 36:1–10. https://doi.org/10.1111/j.1463-6409.2006.00261.x
Watari Y (2019) Roadmap and checklist of invasive species management: learning from the mongoose eradication project on Amami-Oshima. Japanese J Ornithol 68:263–272. https://doi.org/10.3838/jjo.68.263 (in Japanese with English summary)
Watari Y, Takatsuki S, Miyashita T (2008) Effects of exotic mongoose (Herpestes javanicus) on the native fauna of Amami-Oshima Island, southern Japan, estimated by distribution patterns along the historical gradient of mongoose invasion. Biol Invasions 10:7–17. https://doi.org/10.1007/s10530-007-9100-6
Watari Y, Nagata J, Funakoshi K (2011) New detection of a 30-year-old population of introduced mongoose Herpestes auropunctatus on Kyushu Island, Japan. Biol Invasions 13:269–276. https://doi.org/10.1007/s10530-010-9809-5
Watari Y, Nishijima S, Fukasawa M et al (2013) Evaluating the “recovery level” of endangered species without prior information before alien invasion. Ecol Evol 3:4711–4721. https://doi.org/10.1002/ece3.863
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191. https://doi.org/10.1093/genetics/163.3.1177
Wostenberg DJ, Hopken MW, Shiels AB, Piaggio AJ (2019) Using DNA to identify the source of invasive mongooses, Herpestes auropunctatus (Carnivora: Herpestidae) captured on Kaua‘i. Hawaiian Islands Pacific Sci 73:215. https://doi.org/10.2984/73.2.3
Yamada F, Ogura G, Abe S (2015) Herpestes auropunctatus (Hodgson, 1836). In: Ohdachi SD, Ishibashi Y, Iwasa MA, et al. (eds) The wild mammals of Japan, 2nd edn. Shoukadoh Book Sellers, Kyoto, pp 272–274
Yoder AD, Burns MM, Zehr S et al (2003) Single origin of Malagasy Carnivora from an African ancestor. Nat 421:734–737. https://doi.org/10.1038/nature01303
Acknowledgements
We are grateful to Mr. Katsushi Nakata, Dr. Ryoji Fukuhara, Dr. Junco Nagata, Dr. Kimitake Funakoshi, Dr. Shin-Ichiro Kawada, Amami Mongoose Busters, Yambaru Mongoose Busters, and Yambaru Wildlife Center of the Ministry of the Environment, Japan, for providing samples. We also thank Mr. Shintaro Abe and Dr. Fumio Yamada for valuable information and anonymous reviewers for their comments on the draft of this manuscript.
Funding
This research was supported by the Environment Research and Technology Development Fund (JPMEERF20204006) of the Environmental Restoration and Conservation Agency of Japan.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation was performed by Takamichi Jogahara and Yuya Watari. Data collection and analysis were performed by Takuma Sato. The first draft of the manuscript was written by Takuma Sato, and all authors commented on draft versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Communicated by: Jeremy Herman
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sato, T., Watari, Y. & Jogahara, T. Genetic relationships among populations of the small Indian mongoose (Urva auropunctata) introduced in Japan. Mamm Res 68, 177–187 (2023). https://doi.org/10.1007/s13364-023-00672-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13364-023-00672-8