Abstract
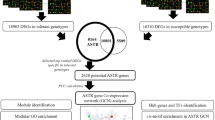
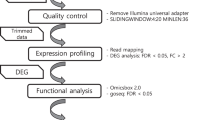
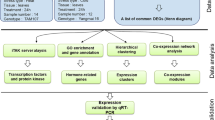
Deteriorating climatic conditions and increasing human population necessitate the development of robust plant varieties resistant to harsh environments. Manipulation of regulatory proteins such as transcription factors (TFs) and RNA-binding proteins (RBPs) would be a beneficial strategy in this regard. Further, understanding the complex interconnections between different classes of regulatory molecules would be essential for the identification of candidate genes/proteins for trait improvement. Most studies to date have analysed the roles of TFs or RBPs individually, in conferring stress resilience. However, it would be important to identify dominant/upstream TFs and RBPs inducing widespread transcriptomic alterations through other regulators (i.e., other TFs/RBPs targeted by the upstream regulators). To this end, the present study employed a transcriptome meta-analysis and computational approaches to obtain a comprehensive overview of regulatory interactions. This work identified dominant TFs and RBPs potentially influencing stress-mediated differential expression of other regulators, which could in turn influence gene expression, and consequently, physiological responses. Twenty transcriptomic studies [related to (i) UV radiation, (ii) wounding, (iii) salinity, (iv) cold, and (v) drought stresses in Arabidopsis thaliana] were analysed for differential gene expression, followed by the identification of differentially expressed TFs and RBPs. Subsequently, other TFs and RBPs which could be influencing these regulators were identified, and their interaction networks and hub nodes were analysed. As a result, an interacting module of Basic Leucine Zipper (bZIP) family TFs as well as Heterogeneous nuclear ribonucleoproteins (hnRNP) and Glycine-rich protein (GRP) family RBPs (among other TFs and RBPs) were shown to potentially influence the stress-induced differential expression of other TFs and RBPs under all the considered stress conditions. Some of the identified hub TFs and RBPs are known to be of major importance in orchestrating stress-induced transcriptomic changes influencing a variety of physiological processes from seed germination to senescence. This study highlighted the gene/protein candidates that could be considered for multiplexed genetic manipulation – a promising approach to develop robust, multi-stress-resilient plant varieties.







Similar content being viewed by others
Data availability
The present study used publicly available microarray data from NCBI-GEO. The considered datasets are mentioned along with references (where available) in the manuscript. The data generated as a part of the present study are available within the manuscript and the supplementary files.
References
Albihlal WS, Gerber AP (2018) Unconventional RNA-binding proteins: an uncharted zone in RNA biology. FEBS Lett 592:2917–2931. https://doi.org/10.1002/1873-3468.13161
Allu AD, Soja AM, Wu A, et al (2014) Salt stress and senescence: Identification of cross-talk regulatory components. J Exp Bot 65:. https://doi.org/10.1093/jxb/eru173
Arae T, Morita K, Imahori R et al (2019) Identification of Arabidopsis CCR4-NOT Complexes with Pumilio RNA-Binding Proteins, APUM5 and APUM2. Plant Cell Physiol 60:2015–2025. https://doi.org/10.1093/pcp/pcz089
Bailey-Serres J, Zhai J, Seki M (2020) The dynamic kaleidoscope of RNA biology in plants. Plant Physiol. 182
Bailey TL, Grant CE (2021) SEA: Simple Enrichment Analysis of motifs. bioRxiv
Barrett T, Wilhite SE, Ledoux P, et al (2013) NCBI GEO: Archive for functional genomics data sets - Update. Nucleic Acids Res 41:. https://doi.org/10.1093/nar/gks1193
Bedre R, Mandadi K (2019) GenFam: A web application and database for gene family-based classification and functional enrichment analysis. Plant Direct 3:. https://doi.org/10.1002/pld3.191
Bhattacharjee A, Jain M (2013) Homeobox genes as potential candidates for crop improvement under abiotic stress. In: Plant Acclimation to Environmental Stress
Bringaud F, Stripecke R, Frech GC, et al (1997) Mitochondrial Glutamate Dehydrogenase from Leishmania tarentolae Is a Guide RNA-Binding Protein . Mol Cell Biol 17:. https://doi.org/10.1128/mcb.17.7.3915
Brown BA, Cloix C, Jiang GH, et al (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci U S A 102:. https://doi.org/10.1073/pnas.0507187102
Chang HC, Tsai MC, Wu SS, Chang IF (2019) Regulation of ABI5 expression by ABF3 during salt stress responses in Arabidopsis thaliana. Bot Stud 60:. https://doi.org/10.1186/s40529-019-0264-z
Chang P, Hsieh HY, Tu SL (2022) The U1 snRNP component RBP45d regulates temperature-responsive flowering in Arabidopsis. Plant Cell 34:834–851. https://doi.org/10.1093/plcell/koab273
Bin CQ, Wang WJ, Zhang Y et al (2022) Abscisic acid-induced cytoplasmic translocation of constitutive photomorphogenic 1 enhances reactive oxygen species accumulation through the HY5-ABI5 pathway to modulate seed germination. Plant Cell Environ 45:1474–1489. https://doi.org/10.1111/pce.14298
Chen LG, Gao Z, Zhao Z et al (2019) BZR1 Family Transcription Factors Function Redundantly and Indispensably in BR Signaling but Exhibit BRI1-Independent Function in Regulating Anther Development in Arabidopsis. Mol Plant 12:1408–1415. https://doi.org/10.1016/j.molp.2019.06.006
Chen YE, Ma J, Wu N et al (2018) The roles of Arabidopsis proteins of Lhcb4, Lhcb5 and Lhcb6 in oxidative stress under natural light conditions. Plant Physiol Biochem 130:267–276. https://doi.org/10.1016/j.plaphy.2018.07.014
Chin CH, Chen SH, Wu HH, et al (2014) cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8:. https://doi.org/10.1186/1752-0509-8-S4-S11
Das M, Haberer G, Panda A, et al (2016) Expression pattern similarities support the prediction of orthologs retaining common functions after gene duplication events. Plant Physiol 171:. https://doi.org/10.1104/pp.15.01207
Díaz-Muñoz MD, Turner M (2018) Uncovering the role of RNA-binding proteins in gene expression in the immune system. Front. Immunol. 9
Dietz KJ, Vogel MO, Viehhauser A (2010) AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245:. https://doi.org/10.1007/s00709-010-0142-8
Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21:972–984. https://doi.org/10.1105/tpc.108.063958
Dro W (2011) Heterodimers of the Arabidopsis Transcription Factors bZIP1 and bZIP53 Reprogram Amino Acid Metabolism during Low Energy Stress. 23:381–395. https://doi.org/10.1105/tpc.110.075390
Foley SW, Gosai SJ, Wang D et al (2017) A Global View of RNA-Protein Interactions Identifies Post-transcriptional Regulators of Root Hair Cell Fate. Dev Cell 41:204-220.e5. https://doi.org/10.1016/j.devcel.2017.03.018
Fu XD, Ares M (2014) Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15
Gangappa SN, Botto JF (2016) The Multifaceted Roles of HY5 in Plant Growth and Development. Mol Plant 9:1353–1365. https://doi.org/10.1016/j.molp.2016.07.002
Ge SX, Jung D, Jung D, Yao R (2020) ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36:. https://doi.org/10.1093/bioinformatics/btz931
Han G, Lu C, Guo J, et al (2020) C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 11
Hu Y, Yu D (2014) BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in arabidopsis. Plant Cell 26:4394–4408. https://doi.org/10.1105/tpc.114.130849
Jaiswal V, Kakkar M, Kumari P, et al (2022) Multifaceted roles of GRAS transcription factors in growth and stress responses in plants. iScience 25:105026. https://doi.org/10.1016/j.isci.2022.105026
Jin H, Li M, Duan S et al (2016) Optimization of light-harvesting pigment improves photosynthetic efficiency. Plant Physiol 172:1720–1731. https://doi.org/10.1104/pp.16.00698
Joshi R, Paul M, Kumar A, Pandey D (2019) Role of calreticulin in biotic and abiotic stress signalling and tolerance mechanisms in plants. Gene 714:144004. https://doi.org/10.1016/j.gene.2019.144004
Khan SA, Li MZ, Wang SM, Yin HJ (2018) Revisiting the role of plant transcription factors in the battle against abiotic stress. Int J Mol Sci 19:. https://doi.org/10.3390/ijms19061634
Kim CY, Bove J, Assmann SM (2008) Overexpression of wound-responsive RNA-binding proteins induces leaf senescence and hypersensitive-like cell death. New Phytol 180:57–70. https://doi.org/10.1111/j.1469-8137.2008.02557.x
Kim JM, To TK, Matsui A, et al (2017) Acetate-mediated novel survival strategy against drought in plants. Nat Plants 3:. https://doi.org/10.1038/nplants.2017.97
Kim JY, Park SJ, Jang B et al (2007) Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J 50:439–451. https://doi.org/10.1111/j.1365-313X.2007.03057.x
Kim S, Kim S, Chang HR, et al (2021) The regulatory impact of RNA-binding proteins on microRNA targeting. Nat Commun 12:. https://doi.org/10.1038/s41467-021-25078-5
Kim Y, Gilmour SJ, Chao L et al (2020) Arabidopsis CAMTA Transcription Factors Regulate Pipecolic Acid Biosynthesis and Priming of Immunity Genes. Mol Plant 13:157–168. https://doi.org/10.1016/j.molp.2019.11.001
Krismer K, Bird MA, Varmeh S et al (2020) Resource Transite : A Computational Motif-Based Analysis Platform That Identifies RNA-Binding Proteins Modulating Changes in Gene Expression ll Transite : A Computational Motif-Based Analysis Platform That Identifies RNA-Binding Proteins Modulating Change. Cell Rep 32:108064. https://doi.org/10.1016/j.celrep.2020.108064
Kupsch C, Ruwe H, Gusewski S et al (2012) Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell 24:4266–4280. https://doi.org/10.1105/tpc.112.103002
Lambermon MHL, Fu Y, Kirk DAW, et al (2002) UBA1 and UBA2, Two Proteins That Interact with UBP1, a Multifunctional Effector of Pre-mRNA Maturation in Plants. Mol Cell Biol 22:. https://doi.org/10.1128/mcb.22.12.4346-4357.2002
Le Roux C, Del Prete S, Boutet-Mercey S, et al (2014) The hnRNP-Q protein LIF2 participates in the plant immune response. PLoS One 9:. https://doi.org/10.1371/journal.pone.0099343
Lee HG, Seo PJ (2015) The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J 82:. https://doi.org/10.1111/tpj.12866
Lee SB, Kim H, Kim RJ, Suh MC (2014) Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep 33:. https://doi.org/10.1007/s00299-014-1636-1
Letunic I, Khedkar S, Bork P (2021) SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res 49:. https://doi.org/10.1093/nar/gkaa937
Li W, Pang S, Lu Z, Jin B (2020) Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 9:1–15. https://doi.org/10.3390/plants9111515
Liu Q, Harberd NP, Fu X (2016) SQUAMOSA Promoter Binding Protein-like Transcription Factors: Targets for Improving Cereal Grain Yield. Mol Plant 9:765–767. https://doi.org/10.1016/j.molp.2016.04.008
Lohani N, Singh MB, Bhalla PL (2022) Biological Parts for Engineering Abiotic Stress Tolerance in Plants. BioDesign Res 2022:. https://doi.org/10.34133/2022/9819314
Mallory MJ, McClory SP, Chatrikhi R et al (2021) Reciprocal regulation of hnRNP C and CELF2 through translation and transcription tunes splicing activity in T cells. Nucleic Acids Res 48:5710–5719. https://doi.org/10.1093/NAR/GKAA295
Marondedze C, Thomas L, Gehring C, Lilley KS (2019) Changes in the Arabidopsis RNA-binding proteome reveal novel stress response mechanisms. BMC Plant Biol 19:. https://doi.org/10.1186/s12870-019-1750-x
Molitor AM, Latrasse D, Zytnicki M et al (2016) The arabidopsis hnRNP-Q protein LIF2 and the PRC1 subunit LHP1 function in concert to regulate the transcription of stress-responsive genes. Plant Cell 28:2197–2211. https://doi.org/10.1105/tpc.16.00244
Mor A, Koh E, Weiner L, et al (2014) Singlet oxygen signatures are detected independent of light or chloroplasts in response to multiple stresses. Plant Physiol 165:. https://doi.org/10.1104/pp.114.236380
Morker KH, Roberts MR (2011) Light exerts multiple levels of influence on the Arabidopsis wound response. Plant, Cell Environ 34:. https://doi.org/10.1111/j.1365-3040.2011.02276.x
Muthusamy M, Kim JH, Kim JA, Lee SI (2021) Plant rna binding proteins as critical modulators in drought, high salinity, heat, and cold stress responses: An updated overview. Int J Mol Sci 22:. https://doi.org/10.3390/ijms22136731
Nguyen CC, Nakaminami K, Matsui A et al (2016) Oligouridylate binding protein 1b plays an integral role in plant heat stress tolerance. Front Plant Sci 7:1–9. https://doi.org/10.3389/fpls.2016.00853
Nguyen CC, Nakaminami K, Matsui A, et al (2017) Overexpression of oligouridylate binding protein 1b results in ABA hypersensitivity. Plant Signal Behav 12:. https://doi.org/10.1080/15592324.2017.1282591
Nishiyama R, Watanabe Y, Leyva-Gonzalez MA, et al (2013) Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc Natl Acad Sci U S A 110:. https://doi.org/10.1073/pnas.1302265110
Nunez-Vazquez R, Desvoyes B, Gutierrez C (2022) Histone variants and modifications during abiotic stress response. Front Plant Sci 13:1–23. https://doi.org/10.3389/fpls.2022.984702
Okuzaki A, Rühle T, Leister D, Schmitz-Linneweber C (2021) The acidic domain of the chloroplast RNA-binding protein CP31A supports cold tolerance in Arabidopsis thaliana. J Exp Bot 72:4904–4914. https://doi.org/10.1093/jxb/erab165
Pan C, Wu X, Markel K, et al (2021) CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat Plants 7:. https://doi.org/10.1038/s41477-021-00953-7
Pandey N, Ranjan A, Pant P, et al (2013) CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics 14:. https://doi.org/10.1186/1471-2164-14-216
Pedrotti L, Weiste C, Nägele T et al (2018) Snf1-RELATED KINASE1-controlled C/S1-bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. Plant Cell 30:495–509. https://doi.org/10.1105/tpc.17.00414
Poudyal RS, Rodionova MV, Kim H et al (2020) Combinatory actions of CP29 phosphorylation by STN7 and stability regulate leaf age-dependent disassembly of photosynthetic complexes. Sci Rep 10:1–10. https://doi.org/10.1038/s41598-020-67213-0
Prall W, Sharma B, Gregory BD (2019) Transcription Is Just the Beginning of Gene Expression Regulation: The Functional Significance of RNA-Binding Proteins to Post-transcriptional Processes in Plants. Plant Cell Physiol 60:1939–1952. https://doi.org/10.1093/pcp/pcz067
Preiss T, Hall AG, Lightowlers RN (1993) Identification of bovine glutamate dehydrogenase as an RNA-binding protein. J Biol Chem 268:. https://doi.org/10.1016/s0021-9258(19)74494-9
Preiss T, Sang AE, Chrzanowska-Lightowlers ZMA, Lightowlers RN (1995) The mRNA-binding protein COLBP is glutamate dehydrogenase. FEBS Lett 367:. https://doi.org/10.1016/0014-5793(95)00569-U
Ramegowda V, Gill US, Sivalingam PN, et al (2017) GBF3 transcription factor imparts drought tolerance in Arabidopsis thaliana. Sci Rep 1–13. https://doi.org/10.1038/s41598-017-09542-1
Rasmussen S, Barah P, Suarez-Rodriguez MC, et al (2013) Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol 161:. https://doi.org/10.1104/pp.112.210773
Ray D, Kazan H, Cook KB, et al (2013) A compendium of RNA-binding motifs for decoding gene regulation. Nature 499:. https://doi.org/10.1038/nature12311
Rivero RM, Mittler R, Blumwald E, Zandalinas SI (2022) Developing climate-resilient crops: improving plant tolerance to stress combination. Plant J 109:. https://doi.org/10.1111/tpj.15483
Shannon P, Markiel A, Ozier O, et al (2003) Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res 13:. https://doi.org/10.1101/gr.1239303
Shi H, Ye T, Zhu JK, Chan Z (2014) Constitutive production of nitric oxide leads to enhanced drought stress resistance and extensive transcriptional reprogramming in Arabidopsis. J Exp Bot 65:. https://doi.org/10.1093/jxb/eru184
Skopelitis DS, Paranychianakis NV, Paschalidis KA et al (2006) Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18:2767–2781. https://doi.org/10.1105/tpc.105.038323
Soboleva TA, Parker BJ, Nekrasov M, et al (2017) A new link between transcriptional initiation and pre-mRNA splicing: The RNA binding histone variant H2A.B. PLoS Genet 13:. https://doi.org/10.1371/journal.pgen.1006633
Sorenson R, Bailey-Serres J (2014) Selective mRNA sequestration by OLIGOURIDYLATEBINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc Natl Acad Sci U S A 111:. https://doi.org/10.1073/pnas.1314851111
Steffen A, Elgner M, Staiger D (2019) Regulation of Flowering Time by the RNA-Binding Proteins AtGRP7 and AtGRP8. Plant Cell Physiol 60:2040–2050. https://doi.org/10.1093/pcp/pcz124
Tian F, Yang DC, Meng YQ, et al (2020) PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res 48:. https://doi.org/10.1093/nar/gkz1020
Walley JW, Kelley DR, Nestorova G, et al (2010) Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol 152:. https://doi.org/10.1104/pp.109.149005
Wang F, Tidei JJ, Polich ED et al (2015) Positive feedback between RNA-binding protein HuD and transcription factor SATB1 promotes neurogenesis. Proc Natl Acad Sci U S A 112:E4995–E5004. https://doi.org/10.1073/pnas.1513780112
Wang L, Cao H, Qian W, et al (2017) Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic arabidopsis. Ann Bot 119:. https://doi.org/10.1093/aob/mcx011
Weiste C, Dro W (2014) The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. https://doi.org/10.1038/ncomms4883
Weiste C, Pedrotti L, Selvanayagam J et al (2017) The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet 13:1–27. https://doi.org/10.1371/journal.pgen.1006607
Wu Z, Zhu D, Lin X et al (2016) RNA binding proteins RZ-1B and RZ-1C play critical roles in regulating pre-mRNA splicing and gene expression during development in arabidopsis. Plant Cell 28:55–73. https://doi.org/10.1105/tpc.15.00949
Xiao Y, Chu L, Zhang Y, et al (2022) HY5 : A Pivotal Regulator of Light-Dependent Development in Higher Plants. 12:1–11. https://doi.org/10.3389/fpls.2021.800989
Yan Y, Gan J, Tao Y et al (2022) RNA-Binding Proteins: The Key Modulator in Stress Granule Formation and Abiotic Stress Response. Front Plant Sci 13:1–18. https://doi.org/10.3389/fpls.2022.882596
Yokoyama M, Hirata KI (2005) New function of calreticulin: Calreticulin-dependent mRNA destabilization. Circ Res 97:961–963. https://doi.org/10.1161/01.RES.0000193564.46466.2a
Yu Y, Qian Y, Jiang M et al (2020) Regulation Mechanisms of Plant Basic Leucine Zippers to Various Abiotic Stresses. Front Plant Sci 11:1–17. https://doi.org/10.3389/fpls.2020.01258
Zhang Y, Xu J, Li R et al (2023) Plants’ Response to Abiotic Stress: Mechanisms and Strategies. Int J Mol Sci 24:10915. https://doi.org/10.3390/ijms241310915
Acknowledgements
Infrastructural facilities provided by St Joseph’s University, Bengaluru, India, are thankfully acknowledged.
Funding
None.
Author information
Authors and Affiliations
Contributions
NMJ: Conceptualization, execution, data-analysis, manuscript writing, and revision.
Corresponding author
Ethics declarations
Competing interests
No relevant financial or non-financial interests exist.
Ethics approval
Not applicable.
Additional information
Communicated by Izabela Pawłowicz.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nishanth, M.J. Transcriptome meta-analysis-based identification of hub transcription factors and RNA-binding proteins potentially orchestrating gene regulatory cascades and crosstalk in response to abiotic stresses in Arabidopsis thaliana. J Appl Genetics 65, 255–269 (2024). https://doi.org/10.1007/s13353-024-00837-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-024-00837-4