Abstract
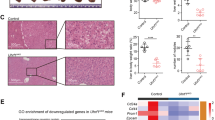
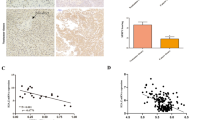
The Wnt pathway is a key regulator of embryonic development and stem cells, and its aberrant activation is associated with human malignancies, most notably hepatocellular carcinoma (HCC). Epigenetic deregulation of the genes encoding the secreted frizzled-related proteins (sFRPs), the Wnt signalling antagonists, has been linked with aberrant hyperactivation of the Wnt signalling in HCC cells; however, the precise underlying mechanism remains elusive. We investigated the methylation profiles of Wnt antagonists in liver samples of different stages of HCC development and liver cancer cell lines and studied the functional impact of aberrant epigenetic silencing of sFRPs on the canonical Wnt pathway and cell viability. We found that the sFRP1 gene encoding the subunit is a frequent target of aberrant DNA hypermethylation and silencing in HCC tumours, whereas other extracellular Wnt antagonists, WIF1 and Dkk3, exhibited no methylation in tumour cells, consistent with the notion that aberrant methylation events in cancer cells are non-randomly distributed among the genes and that there is a strong preference for hypermethylation of specific genes in HCC. In addition, by comparing sFRP1 methylation status in HCC tumours with normal, cirrhotic and chronic hepatitis liver tissues, we identified sFRP1 gene as a potential early marker of HCC. The restoration of sFRP1 expression in cancer cells by ectopic expression inhibited Wnt activity accompanied with destabilization of β-catenin and downregulation of c-Myc and cyclin D1, the known downstream targets of Wnt pathway. Importantly, restoring sFRP1 levels in cancer cells inhibited cell growth and induced apoptotic cell death. This study supports the critical role for sFRP1 silencing in hepatocellular carcinoma and reinforces the importance of the Wnt antagonists in preventing oncogenic stabilization of β-catenin and chronic activation of the canonical Wnt pathway, suggesting that sFRP1 may be an attractive target for early cancer detection and therapeutic intervention.




Similar content being viewed by others
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- HBV:
-
Hepatitis B virus
- HCV:
-
Hepatitis C virus
- DMEM:
-
Dulbecco’s minimum essential medium
- RT-QPCR:
-
Real-time quantitative transcription PCR
- sFRPs:
-
Secreted frizzled-related proteins
- WIF1:
-
Wnt inhibitory factor 1
- Dkk3:
-
Dickkopf 3
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
References
Globocan (2008) International Agency for Research on Cancer, Lyon, France
Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6(2):107–16.
Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92.
Herceg Z, Paliwal A. Epigenetic mechanisms in hepatocellular carcinoma: how environmental factors influence the epigenome. Mutat Res. 2011;727(3):55–61.
Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14(15):1837–51.
de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O, et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci U S A. 1998;95(15):8847–51.
Miyoshi Y, Iwao K, Nagasawa Y, Aihara T, Sasaki Y, Imaoka S, et al. Activation of the beta-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res. 1998;58(12):2524–7.
Huang H, Fujii H, Sankila A, Mahler-Araujo BM, Matsuda M, Cathomas G, et al. Beta-catenin mutations are frequent in human hepatocellular carcinomas associated with hepatitis C virus infection. Am J Pathol. 1999;155(6):1795–801.
Chien AJ, Conrad WH, Moon RT. A Wnt survival guide: from flies to human disease. J Invest Dermatol. 2009;129(7):1614–27.
Jones SE, Jomary C. Secreted frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24(9):811–20.
Chong JM, Uren A, Rubin JS, Speicher DW. Disulfide bond assignments of secreted frizzled-related protein-1 provide insights about frizzled homology and netrin modules. J Biol Chem. 2002;277(7):5134–44.
Caldwell GM, Jones C, Gensberg K, Jan S, Hardy RG, Byrd P, et al. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 2004;64(3):883–8.
Tanaka J, Watanabe T, Kanazawa T, Tada T, Kazama Y, Tanaka T, et al. Silencing of secreted frizzled-related protein genes in MSI colorectal carcinogenesis. Hepatogastroenterology. 2008;55(85):1265–8.
Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12(44):7113–7.
Zou H, Molina JR, Harrington JJ, Osborn NK, Klatt KK, Romero Y, et al. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett’s esophagus. Int J Cancer. 2005;116(4):584–91.
Clement G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J. Alterations of the Wnt signaling pathway during the neoplastic progression of Barrett’s esophagus. Oncogene. 2006;25(21):3084–92.
Stoehr R, Wissmann C, Suzuki H, Knuechel R, Krieg RC, Klopocki E, et al. Deletions of chromosome 8p and loss of sFRP1 expression are progression markers of papillary bladder cancer. Lab Invest. 2004;84(4):465–78.
Urakami S, Shiina H, Enokida H, Kawakami T, Kawamoto K, Hirata H, et al. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res. 2006;12(7 Pt 1):2109–16.
Nojima M, Suzuki H, Toyota M, Watanabe Y, Maruyama R, Sasaki S, et al. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene. 2007;26(32):4699–713.
Zhao CH, Bu XM, Zhang N. Hypermethylation and aberrant expression of Wnt antagonist secreted frizzled-related protein 1 in gastric cancer. World J Gastroenterol. 2007;13(15):2214–7.
Morris MR, Ricketts C, Gentle D, Abdulrahman M, Clarke N, Brown M, et al. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene. 2010;29(14):2104–17.
Costa VL, Henrique R, Ribeiro FR, Carvalho JR, Oliveira J, Lobo F, et al. Epigenetic regulation of Wnt signaling pathway in urological cancer. Epigenetics. 2010;5(4):343–51.
Fukui T, Kondo M, Ito G, Maeda O, Sato N, Yoshioka H, et al. Transcriptional silencing of secreted frizzled related protein 1 (SFRP 1) by promoter hypermethylation in non-small-cell lung cancer. Oncogene. 2005;24(41):6323–7.
Tang M, Torres-Lanzas J, Lopez-Rios F, Esteller M, Sanchez-Cespedes M. Wnt signaling promoter hypermethylation distinguishes lung primary adenocarcinomas from colorectal metastasis to the lung. Int J Cancer. 2006;119(11):2603–6.
Shih YL, Hsieh CB, Lai HC, Yan MD, Hsieh TY, Chao YC, et al. SFRP1 suppressed hepatoma cells growth through Wnt canonical signaling pathway. Int J Cancer. 2007;121(5):1028–35.
Takagi H, Sasaki S, Suzuki H, Toyota M, Maruyama R, Nojima M, et al. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol. 2008;43(5):378–89.
Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8(1):91–9.
Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275(5307):1784–7.
Finkbeiner MG, Sawan C, Ouzounova M, Murr R, Herceg Z. HAT cofactor TRRAP mediates beta-catenin ubiquitination on the chromatin and the regulation of the canonical Wnt pathway. Cell Cycle. 2008;7(24):3908–14.
Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000;19(8):1839–50.
Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16(13):3797–804.
Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17(3):330–9.
Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22(2):91–103.
Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–56.
Agrawal A, Murphy RF, Agrawal DK. DNA methylation in breast and colorectal cancers. Mod Pathol. 2007;20(7):711–21.
Acknowledgement
P. Kaur and S. Mani are supported by a post-doctoral fellowship from the International Agency for Research on Cancer (IARC), Lyon, France. This work in the IARC Epigenetics Group is supported by grants from l’Agence Nationale de Recherhe Contre le Sida et Hépatites Virales (ANRS, France); the Association pour la Recherche sur le Cancer (ARC, France); la Ligue Nationale (Française) Contre le Cancer (France); and the Swiss Bridge Award.
Financial disclosure
This work was supported by la Ligue Nationale (Française) Contre le Cancer, l’Association pour le Recherche Contre le Cancer (l’ARC), l’Agence Nationale de Recherche Contre le Sida et Hépatites Virales (ANRS, France) and Swiss Bridge Award. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 71 kb)
Rights and permissions
About this article
Cite this article
Kaur, P., Mani, S., Cros, MP. et al. Epigenetic silencing of sFRP1 activates the canonical Wnt pathway and contributes to increased cell growth and proliferation in hepatocellular carcinoma. Tumor Biol. 33, 325–336 (2012). https://doi.org/10.1007/s13277-012-0331-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-012-0331-5