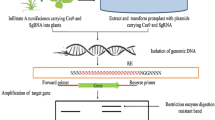
Abstract
The exponentially increasing population poses a serious threat to global food security. Concurrently, the climate change condition also limits crop productivity by enhancing the effect of biotic and abiotic stressors. The traditional crop improvement programs are not enough to meet the food and nutritional requirements of such a progressive population. Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) based genome editing tool adopted from bacterial adaptive immune system against invading foreign DNA has demonstrated its tremendous potential in the sector of crop improvement. CRISPR/Cas-mediated targeting can activate, repress, or completely abolish the gene function. CRISPR/Cas-mediated interventions can produce biofortified crops by targeting negative regulators or activating positive regulators for nutrients. Thus, it can address nutritional security concerns. The advancement in CRISPR/Cas-mediated genome editing, encompassing base and prime editing has paved the way to modify an organism’s genome in a predictable and precise manner. The use of morphogenetic regulators can omit the problem of tissue culture stages, which is one of the major bottlenecks in plant genome editing. CRISPR/Cas-based genome editing has been performed in many crop plants to induce biotic and abiotic stress tolerance, increase quality and nutritional values, enhance productivity, and prevent post-harvest losses. In this review article, we summarize the progress, challenges opportunities and regulatory landscape of genome editing for the improvement of various traits in crop plants.
Graphical abstract




Similar content being viewed by others
Data availability
Data sharing does not apply to this article as no datasets were generated or analyzed during the current study.
References
Abdallah NA, Elsharawy H, Abulela HA, Thilmony R, Abdelhadi AA, Elarabi NI. Multiplex CRISPR/Cas9-mediated genome editing to address drought tolerance in wheat. GM Crops Food. 2022;6:1–17. https://doi.org/10.1080/21645698.2022.2120313.
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. RNA targeting with CRISPR-Cas13. Nature. 2017;550(7675):280–4. https://doi.org/10.1038/nature24049.
Acharya S, Mishra A, Paul D, Ansari AH, Azhar M, Kumar M, Rauthan R, Sharma N, Aich M, Sinha D, Sharma S, Jain S, Ray A, Jain S, Ramalingam S, Maiti S, Chakraborty D. Francisella novicida Cas9 interrogates genomic DNA with very high specificity and can be used for mammalian genome editing. Proc Natl Acad Sci USA. 2019;116(42):20959–68. https://doi.org/10.1073/pnas.1818461116.
Akama K, Akter N, Endo H, Kanesaki M, Endo M, Toki S. An in vivo targeted deletion of the calmodulin-binding domain from rice glutamate decarboxylase 3 (OsGAD3) increases γ-aminobutyric acid content in grains. Rice. 2020;13(1):20. https://doi.org/10.1186/s12284-020-00380-w.
Alam MS, Kong J, Tao R, Ahmed T, Alamin M, Alotaibi SS, Abdelsalam NR, Xu JH. CRISPR/Cas9 mediated knockout of the OsbHLH024 transcription factor improves salt stress resistance in Rice (Oryza sativa L.). Plants (Basel). 2022;11(9):1184. https://doi.org/10.3390/plants11091184.
Alfatih A, Wu J, Jan SU, Zhang ZS, Xia JQ, Xiang CB. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 2020;43(11):2743–54. https://doi.org/10.1111/pce.13856.
Ali Z, Abulfaraj A, Idris A, Ali S, Tashkandi M, Mahfouz MM. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015;16:238. https://doi.org/10.1186/s13059-015-0799-6.
Ali Z, Ali S, Tashkandi M, Zaidi SS, Mahfouz MM. CRISPR/Cas9-mediated immunity to geminiviruses: differential interference and evasion. Sci Rep. 2016;6:26912. https://doi.org/10.1038/srep26912.
Anzalone AV, Gao XD, Podracky CJ, Nelson AT, Koblan LW, Raguram A, Levy JM, Mercer JAM, Liu DR. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat Biotechnol. 2022;40(5):731–40. https://doi.org/10.1038/s41587-021-01133-w.
Ashokkumar S, Jaganathan D, Ramanathan V, Rahman H, Palaniswamy R, Kambale R, Muthurajan R. Creation of novel alleles of fragrance gene OsBADH2 in rice through CRISPR/Cas9 mediated gene editing. PLoS ONE. 2020;15(8):e0237018. https://doi.org/10.1371/journal.pone.0237018.
Awasthi R, Bhandari K, Nayyar H. Temperature stress and redox homeostasis in agricultural crops. Front Environ Sci. 2015;3:11. https://doi.org/10.3389/fenvs.2015.00011.
Awasthi P, Khan S, Lakhani H, Chaturvedi S, Shivani KN, Singh J, Kesarwani AK, Tiwari S. Transgene-free genome editing supports CCD4 role as a negative regulator of β-carotene in banana. J Exp Bot. 2022;73:erac042. https://doi.org/10.1093/jxb/erac042.
Badhan S, Ball AS, Mantri N. First report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts. Int J Mol Sci. 2021;22(1):396. https://doi.org/10.3390/ijms22010396.
Baeg GJ, Kim SH, Choi DM, Tripathi S, Han YJ, Kim JI. CRISPR/Cas9-mediated mutation of 5-oxoprolinase gene confers resistance to sulfonamide compounds in Arabidopsis. Plant Biotechnol Rep. 2021;15:753–64. https://doi.org/10.1007/s11816-021-00718-w.
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF. DNA replicons for plant genome engineering. Plant Cell. 2014;26(1):151–63. https://doi.org/10.1105/tpc.113.119792.
Baltes NJ, Hummel AW, Konecna E, Cegan R, Bruns AN, Bisaro DM, Voytas DF. Conferring resistance to geminiviruses with the CRISPR/Cas prokaryotic immune system. Nat Plants. 2015;1(10):15145. https://doi.org/10.1038/nplants.2015.145.
Ben Shlush I, Samach A, Melamed-Bessudo C, Ben-Tov D, Dahan-Meir T, Filler-Hayut S, Levy AA. CRISPR/Cas9 induced somatic recombination at the CRTISO locus in tomato. Genes (Basel). 2020;12(1):59. https://doi.org/10.3390/genes12010059.
Beracochea V, Stritzler M, Radonic L, Bottero E, Jozefkowicz C, Darqui F, Ayub N, Bilbao ML, Soto G. CRISPR/Cas9-mediated knockout of SPL13 radically increases lettuce yield. Plant Cell Rep. 2023;42(3):645–7. https://doi.org/10.1007/s00299-022-02952-0.
Bertier LD, Ron M, Huo H, Bradford KJ, Britt AB, Michelmore RW. High-resolution analysis of the efficiency, heritability, and editing outcomes of CRISPR/Cas9-induced modifications of NCED4 in lettuce (Lactuca sativa). G3 Genes Genomes Genet. 2018;8(5):1513–21. https://doi.org/10.1534/g3.117.300396.
Beying N, Schmidt C, Pacher M, Houben A, Puchta H. CRISPR-Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat Plants. 2020;6(6):638–45. https://doi.org/10.1038/s41477-020-0663-x.
Bharathkumar N, Sunil A, Meera P, Aksah S, Kannan M, Saravanan KM, Anand T. CRISPR/Cas-based modifications for therapeutic applications: a review. Mol Biotechnol. 2022;64(4):355–72. https://doi.org/10.1007/s12033-021-00422-8.
Bouzroud S, Gasparini K, Hu G, Barbosa MAM, Rosa BL, Fahr M, Bendaou N, Bouzayen M, Zsögön A, Smouni A, Zouine M. Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes (Basel). 2020;11(3):272. https://doi.org/10.3390/genes11030272.
Buchholzer M, Frommer WB. An increasing number of countries regulate genome editing in crops. New Phytol. 2023;237(1):12–5. https://doi.org/10.1111/nph.18333.
Budhagatapalli N, Schedel S, Gurushidze M, Pencs S, Hiekel S, Rutten T, Kusch S, Morbitzer R, Lahaye T, Panstruga R, Kumlehn J, Hensel G. A simple test for the cleavage activity of customized endonucleases in plants. Plant Methods. 2016;12:18. https://doi.org/10.1186/s13007-016-0118-6.
Cai Q, Guo D, Cao Y, Li Y, Ma R, Liu W. Application of CRISPR/CasΦ2 system for genome editing in plants. Int J Mol Sci. 2022;23(10):5755. https://doi.org/10.3390/ijms23105755.
Calyxt, I.: First commercial sale of calyxt high oleic soybean oil on the U.S. Market. https://calyxt.com/first-commercial-sale-of-calyxt-high-oleic-soybean-oil-on-the-u-s-market/. (2019). Accessed 01 July 2023.
Čermák T, Baltes NJ, Čegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. https://doi.org/10.1186/s13059-015-0796-9.
Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol. 2016;17(7):1140–53. https://doi.org/10.1111/mpp.12375.
Chaudhary R, Singh S, Kaur K, Tiwari S. Genome-wide identification and expression profiling of WUSCHEL-related homeobox (WOX) genes confer their roles in somatic embryogenesis, growth and abiotic stresses in banana. 3 Biotech. 2022;12(11):321. https://doi.org/10.1007/s13205-022-03387-w.
Che P, Wu E, Simon MK, Anand A, Lowe K, Gao H, Sigmund AL, Yang M, Albertsen MC, Gordon-Kamm W, Jones TJ. Wuschel2 enables highly efficient CRISPR/Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. Commun Biol. 2022;5(1):344. https://doi.org/10.1038/s42003-022-03308-w.
Che J, Yamaji N, Ma JF. Role of a vacuolar iron transporter OsVIT2 in the distribution of iron to rice grains. New Phytol. 2021;230(3):1049–62. https://doi.org/10.1111/nph.17219.
Chen H, Su Z, Tian B, Liu Y, Pang Y, Kavetskyi V, Trick HN, Bai G. Development and optimization of a Barley stripe mosaic virus-mediated gene editing system to improve Fusarium head blight resistance in wheat. Plant Biotechnol J. 2022;20(6):1018–20. https://doi.org/10.1111/pbi.13819.
Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol. 2019;70:667–97. https://doi.org/10.1146/annurev-arplant-050718-100049.
Chen S, Zhang N, Zhou G, Hussain S, Ahmed S, Tian H, Wang S. Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 2021;21(1):137. https://doi.org/10.1186/s12870-021-02907-9.
Chu C, Huang R, Liu L, Tang G, Xiao J, Yoo H, Yuan M. The rice heavy-metal transporter OsNRAMP1 regulates disease resistance by modulating ROS homoeostasis. Plant Cell Environ. 2022;45(4):1109–26. https://doi.org/10.1111/pce.14263.
Cui Y, Jiang N, Xu Z, Xu Q. Heterotrimeric G protein are involved in the regulation of multiple agronomic traits and stress tolerance in rice. BMC Plant Biol. 2020;20(1):90. https://doi.org/10.1186/s12870-020-2289-6.
Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, Bocobza S, Czosnek H, Aharoni A, Levy AA. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR-Cas9 system. Plant J. 2018;95(1):5–16. https://doi.org/10.1111/tpj.13932.
Debernardi JM, Tricoli DM, Ercoli MF, Hayta S, Ronald P, Palatnik JF, Dubcovsky J. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat Biotechnol. 2020;38(11):1274–9. https://doi.org/10.1038/s41587-020-0703-0.
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–7. https://doi.org/10.1038/nature09886.
Do PT, Nguyen CX, Bui HT, Tran LTN, Stacey G, Gillman JD, Zhang ZJ, Stacey MG. Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol. 2019;19(1):311. https://doi.org/10.1186/s12870-019-1906-8.
Dong OX, Yu S, Jain R, Zhang N, Duong PQ, Butler C, Li Y, Lipzen A, Martin JA, Barry KW, Schmutz J, Tian L, Ronald PC. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR/Cas9. Nat Commun. 2020;11(1):1178. https://doi.org/10.1038/s41467-020-14981-y.
Entine J, Felipe MSS, Groenewald JH, Kershen DL, Lema M, McHughen A, Nepomuceno AL, Ohsawa R, Ordonio RL, Parrott WA, Quemada H, Ramage C, Slamet-Loedin I, Smyth SJ, Wray-Cahen D. Regulatory approaches for genome edited agricultural plants in select countries and jurisdictions around the world. Transgenic Res. 2021;30(4):551–84. https://doi.org/10.1007/s11248-021-00257-8.
FAO. The future of food and agriculture—trends and challenges. Rome: FAO. (2017).
Gao H, Gadlage MJ, Lafitte HR, Lenderts B, Yang M, Schroder M, Farrell J, Snopek K, Peterson D, Feigenbutz L, Jones S, St Clair G, Rahe M, Sanyour-Doyel N, Peng C, Wang L, Young JK, Beatty M, Dahlke B, Hazebroek J, Greene TW, Cigan AM, Chilcoat ND, Meeley RB. Superior field performance of waxy corn engineered using CRISPR/Cas9. Nat Biotechnol. 2020;38(5):579–81. https://doi.org/10.1038/s41587-020-0444-0.
Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003;67(1):16–37. https://doi.org/10.1128/MMBR.67.1.16-37.2003.
Gong Z, Xiong L, Shi H, Yang S, Herrera-Estrella LR, Xu G, Chao DY, Li J, Wang PY, Qin F, Li J, Ding Y, Shi Y, Wang Y, Yang Y, Guo Y, Zhu JK. Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci. 2020;63(5):635–74. https://doi.org/10.1007/s11427-020-1683-x.
Gordon-Kamm B, Sardesai N, Arling M, Lowe K, Hoerster G, Betts S, Jones AT. Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants (Basel). 2019;8(2):38. https://doi.org/10.3390/plants8020038.
He X, Luo X, Wang T, Liu S, Zhang X, Zhu L. GhHB12 negatively regulates abiotic stress tolerance in Arabidopsis and cotton. Environ Exp Bot. 2020;176:104087. https://doi.org/10.1016/j.envexpbot.2020.104087.
Hernandes-Lopes J, Pinto MS, Vieira LR, Monteiro PB, Gerasimova SV, Nonato JVA, Bruno MHF, Vikhorev A, Rausch-Fernandes F, Gerhardt IR, Pauwels L, Arruda P, Dante RA, Yassitepe JECT. Enabling genome editing in tropical maize lines through an improved, morphogenic regulator-assisted transformation protocol. Front Genome Ed. 2023;5:1241035. https://doi.org/10.3389/fgeed.2023.1241035.
Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci USA. 2014;111(18):6618–23. https://doi.org/10.1073/pnas.1405079111.
Hofvander P, Andreasson E, Andersson M. Potato trait development going fast-forward with genome editing. Trends Genet. 2022;38(3):218–21. https://doi.org/10.1016/j.tig.2021.10.004.
Hong Y, Meng J, He X, Zhang Y, Liu Y, Zhang C, Qi H, Luan Y. Editing miR482b and miR482c simultaneously by CRISPR/Cas9 enhanced tomato resistance to phytophthora infestans. Phytopathology. 2021;111(6):1008–16. https://doi.org/10.1094/phyto-08-20-0360-r.
Hu Z, Li J, Ding S, Cheng F, Li X, Jiang Y, Yu J, Foyer CH, Shi K. The protein kinase CPK28 phosphorylates ascorbate peroxidase and enhances thermotolerance in tomato. Plant Physiol. 2021;186(2):1302–17. https://doi.org/10.1093/plphys/kiab120.
Hu C, Sheng O, Deng G, He W, Dong T, Yang Q, Dou T, Li C, Gao H, Liu S, Yi G, Bi F. CRISPR/Cas9-mediated genome editing of MaACO1 (aminocyclopropane-1-carboxylate oxidase 1) promotes the shelf life of banana fruit. Plant Biotechnol J. 2021;19(4):654–6. https://doi.org/10.1111/pbi.13534.
Hua K, Jiang Y, Tao X, Zhu JK. Precision genome engineering in rice using prime editing system. Plant Biotechnol J. 2020;18(11):2167–9. https://doi.org/10.1111/pbi.13395.
Hua K, Tao X, Liang W, Zhang Z, Gou R, Zhu JK. Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotechnol J. 2020;18(3):770–8. https://doi.org/10.1111/pbi.13244.
Hua K, Tao X, Yuan F, Wang D, Zhu JK. Precise A·T to G·C base editing in the rice genome. Mol Plant. 2018;11(4):627–30. https://doi.org/10.1016/j.molp.2018.02.007.
Huang X, Zeng X, Li J, Zhao D. Construction and analysis of tify1a and tify1b mutants in rice (Oryza sativa) based on CRISPR/Cas9 technology. J Agric Biotechnol. 2017;25(6):1003–12.
Hussain B, Lucas SJ, Budak H. CRISPR/Cas9 in plants: at play in the genome and at work for crop improvement. Brief Funct Genomics. 2018;17(5):319–28. https://doi.org/10.1093/bfgp/ely016.
Jacobs TB, Zhang N, Patel D, Martin GB. Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol. 2017;174(4):2023–37. https://doi.org/10.1104/pp.17.00489.
Javaid D, Ganie SY, Hajam YA, Reshi MS. CRISPR/Cas9 system: a reliable and facile genome editing tool in modern biology. Mol Biol Rep. 2022;49(12):12133–50. https://doi.org/10.1007/s11033-022-07880-6.
Ji X, Zhang H, Zhang Y, Wang Y, Gao C. Establishing a CRISPR/Cas-like immune system conferring DNA virus resistance in plants. Nat Plants. 2015;1:15144. https://doi.org/10.1038/nplants.2015.144.
Jia H, Zhang Y, Orbović V, Xu J, White FF, Jones JB, Wang N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol J. 2017;15(7):817–23. https://doi.org/10.1111/pbi.12677.
Jiang YY, Chai YP, Lu MH, Han XL, Lin Q, Zhang Y, Zhang Q, Zhou Y, Wang XC, Gao C, Chen QJ. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 2020;21(1):257. https://doi.org/10.1186/s13059-020-02170-5.
Jiang WZ, Henry IM, Lynagh PG, Comai L, Cahoon EB, Weeks DP. Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol J. 2017;15(5):648–57. https://doi.org/10.1111/pbi.12663.
Jin S, Zong Y, Gao Q, Zhu Z, Wang Y, Qin P, Liang C, Wang D, Qiu JL, Zhang F, Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364(6437):292–5. https://doi.org/10.1126/science.aaw7166. (Epub 2019 Feb 28 PMID: 30819931).
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. https://doi.org/10.1126/science.1225829.
Juma BS, Mukami A, Mweu C, Ngugi MP, Mbinda W. Targeted mutagenesis of the CYP79D1 gene via CRISPR/Cas9-mediated genome editing results in lower levels of cyanide in cassava. Front Plant Sci. 2022;26(13):1009860. https://doi.org/10.3389/fpls.2022.1009860.PMID:36388608;PMCID:PMC9644188.
Kaur N, Alok A, Shivani KP, Kaur N, Awasthi P, Chaturvedi S, Pandey P, Pandey A, Pandey AK, Tiwari S. CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit. Metab Eng. 2020;59:76–86. https://doi.org/10.1016/j.ymben.2020.01.008.
Kim D, Alptekin B, Budak H. CRISPR/Cas9 genome editing in wheat. Funct Integr Genomics. 2018;18(1):31–41. https://doi.org/10.1007/s10142-017-0572-x.
Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to FokI cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):1156–60. https://doi.org/10.1073/pnas.93.3.1156.
Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun. 2017;8:14406. https://doi.org/10.1038/ncomms14406.
Kodackattumannil P, Lekshmi G, Kottackal M, Sasi S, Krishnan S, Al Senaani S, Amiri KMA. Hidden pleiotropy of agronomic traits uncovered by CRISPR-Cas9 mutagenesis of the tyrosinase CuA-binding domain of the polyphenol oxidase 2 of eggplant. Plant Cell Rep. 2023;42(4):825–8. https://doi.org/10.1007/s00299-023-02987-x.
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–4. https://doi.org/10.1038/nature17946.
Kong J, Martin-Ortigosa S, Finer J, Orchard N, Gunadi A, Batts LA, Thakare D, Rush B, Schmitz O, Stuiver M, Olhoft P, Pacheco-Villalobos D. Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocotspecies. Front Plant Sci. 2020;11:572319. https://doi.org/10.3389/fpls.2020.572319.
Kuang Y, Li S, Ren B, Yan F, Spetz C, Li X, Zhou X, Zhou H. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol Plant. 2020;13(4):565–72. https://doi.org/10.1016/j.molp.2020.01.010.
Kumar S. Abiotic stresses and their effects on plant growth, yield and nutritional quality of agricultural produce. Int J Food Sci Agric. 2020;4:367–78. https://doi.org/10.26855/ijfsa.2020.12.002.
Kurt IC, Zhou R, Iyer S, Garcia SP, Miller BR, Langner LM, Grünewald J, Joung JK. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol. 2021;39(1):41–6. https://doi.org/10.1038/s41587-020-0609-x.
Kwabena Osei M, Danquah A, Adu-Dapaah H, Danquah E, Blay E, Massoudi M, Maxwell D. Marker assisted backcrossing of alcobaca gene into two elite tomato breeding lines. Front Hortic. 2022;22(1):1024042.
Laura M, Forti C, Barberini S, Ciorba R, Mascarello C, Giovannini A, Pistelli L, Pieracci Y, Lanteri AP, Ronca A, et al. Highly efficient CRISPR/Cas9 mediated gene editing in Ocimum basilicum ‘FT Italiko’ to induce resistance to Peronospora belbahrii. Plants. 2023;12(13):2395. https://doi.org/10.3390/plants12132395.
Lei J, Dai P, Li Y, Zhang W, Zhou G, Liu C, Liu X. Heritable gene editing using FT mobile guide RNAs and DNA viruses. Plant Methods. 2021;17(1):1–20. https://doi.org/10.1186/s13007-021-00719-4.
Li X, Hu D, Cai L, Wang H, Liu X, Du H, Yang Z, Zhang H, Hu Z, Huang F, Kan G, Kong F, Liu B, Yu D, Wang H. CALCIUM-DEPENDENT PROTEIN KINASE38 regulates flowering time and common cutworm resistance in soybean. Plant Physiol. 2022;190(1):480–99. https://doi.org/10.1093/plphys/kiac260.
Li R, Li R, Li X, Fu D, Zhu B, Tian H, Luo Y, Zhu H. Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol J. 2018;16(2):415–27. https://doi.org/10.1111/pbi.12781.
Li Y, Liang J, Deng B, Jiang Y, Zhu J, Chen L, Li M, Li J. Applications and prospects of CRISPR/Cas9-mediated base editing in plant breeding. Curr Issues Mol Biol. 2023;45(2):918–35. https://doi.org/10.3390/cimb45020059.
Li J, Meng X, Zong Y, Chen K, Zhang H, Liu J, Li J, Gao C. Gene replacements and insertions in rice by intron targeting using CRISPR/Cas9. Nat Plants. 2016;2:16139. https://doi.org/10.1038/nplants.2016.139.
Li J, Pan W, Zhang S, Ma G, Li A, Zhang H, Liu L. A rapid and highly efficient sorghum transformation strategy using GRF4-GIF1/ternary vector system. Plant J. 2023;28(9):1998–2015. https://doi.org/10.1105/tpc.16.00124.
Li J, Sun Y, Du J, Zhao Y, Xia L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant. 2017;10(3):526–9. https://doi.org/10.1016/j.molp.2016.12.001.
Li X, Wang Y, Chen S, Tian H, Fu D, Zhu B, Luo Y, Zhu H. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front Plant Sci. 2018;9:559. https://doi.org/10.3389/fpls.2018.00559.
Li R, Wang L, Chen L, Yu W, Zhang S, Sheng J, Shen L. CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 2019;19(1):38. https://doi.org/10.1186/s12870-018-1627-4.
Li G, Zhang Y, Dailey M, Qi Y. Hs1Cas12a and Ev1Cas12a confer efficient genome editing in plants. Front Genome Ed. 2023;5:1251903. https://doi.org/10.3389/fgeed.2023.1251903.
Li J, Zhang H, Si X, Tian Y, Chen K, Liu J, Chen H, Gao C. Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J Genet Genomics. 2017;44(9):465–8. https://doi.org/10.1016/j.jgg.2017.02.002.
Li R, Zhang L, Wang L, Chen L, Zhao R, Sheng J, Shen L. Reduction of tomato-plant chilling tolerance by CRISPR/Cas9-mediated SlCBF1 mutagenesis. J Agric Food Chem. 2018;66(34):9042–51. https://doi.org/10.1021/acs.jafc.8b02177.
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018;19(1):59. https://doi.org/10.1186/s13059-018-1443-z.
Liao S, Qin X, Luo L, Han Y, Wang X, Usman B, Nawaz G, Zha N, Liu Y, Li R. CRISPR/Cas9-induced mutagenesis of Semi-rolled leaf1, 2 Confers curled leaf phenotype and drought tolerance by influencing protein expression patterns and ROS scavenging in rice (Oryza sativa L.). Agronomy. 2019;9(11):728. https://doi.org/10.3390/agronomy9110728.
Lin Q, Jin S, Zong Y, Yu H, Zhu Z, Liu G, Kou L, Wang Y, Qiu JL, Li J, Gao C. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat Biotechnol. 2021;39(8):923–7. https://doi.org/10.1038/s41587-021-00868-w.
Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone AV, Raguram A, Doman JL, Liu DR, Gao C. Prime genome editing in rice and wheat. Nat Biotechnol. 2020;38(5):582–5. https://doi.org/10.1038/s41587-020-0455-x.
Liu L, Chen P, Wang M, Li X, Wang J, Yin M, Wang Y. C2c1-sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol Cell. 2017;65(2):310–22. https://doi.org/10.1016/j.molcel.2016.11.040.
Liu Y, Chen Z, Zhang C, Guo J, Liu Q, Yin Y, Hu Y, Xia H, Li B, Sun X, Li Y, Liu X. Gene editing of ZmGA20ox3 improves plant architecture and drought tolerance in maize. Plant Cell Rep. 2023;43(1):18. https://doi.org/10.1007/s00299-023-03090-x.
Liu X, Wu D, Shan T, Xu S, Qin R, Li H, Negm M, Wu D, Li J. The trihelix transcription factor OsGTγ-2 is involved in adaption to salt stress in rice. Plant Mol Biol. 2020;103(4–5):545–60. https://doi.org/10.1007/s11103-020-01010-1.
Liu D, Xuan S, Prichard LE, Donahue LI, Pan C, Nagalakshmi U, Ellison EE, Starker CG, Dinesh-Kumar SP, Qi Y, Voytas DF. Heritable base-editing in Arabidopsis using RNA viral vectors. Plant Physiol. 2022;189(4):1920–4. https://doi.org/10.1093/plphys/kiac206.
Liu T, Zeng D, Zheng Z, Lin Z, Xue Y, Li T, Xie X, Ma G, Liu YG, Zhu Q. The ScCas9++ variant expands the CRISPR toolbox for genome editing in plants. J Integr Plant Biol. 2021;63(9):1611–9. https://doi.org/10.1111/jipb.13164.
Liu Y, Zhang L, Li C, Yang Y, Duan Y, Yang Y, Sun X. Establishment of Agrobacterium-mediated genetic transformation and application of CRISPR/Cas9 genome-editing system to Brassica rapa var. rapa. Plant Methods. 2022;18(1):98. https://doi.org/10.1186/s13007-022-00931-w.
Liu L, Zhang J, Xu J, Li Y, Guo L, Wang Z, Zhang X, Zhao B, Guo YD, Zhang N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020;301:110683. https://doi.org/10.1016/j.plantsci.2020.110683.
Lou D, Wang H, Liang G, Yu D. OsSAPK2 Confers abscisic acid sensitivity and tolerance to drought stress in rice. Front Plant Sci. 2017;8:993. https://doi.org/10.3389/fpls.2017.00993.
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang L, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao Z, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng P, Bidney D, Falco C, Register J, Zhao ZY, Xu D, Jones T, Gordon-Kamm W. Morphogenic regulators baby boom and wuschel improve monocot transformation. Plant Cell. 2016;28(9):1998–2015. https://doi.org/10.1105/tpc.16.00124.
Lu Y, Tian Y, Shen R, Yao Q, Wang M, Chen M, Dong J, Zhang T, Li F, Lei M, Zhu JK. Targeted, efficient sequence insertion and replacement in rice. Nat Biotechnol. 2020;38(12):1402–7. https://doi.org/10.1038/s41587-020-0581-5.
Lu Y, Zhu JK. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol Plant. 2017;10(3):523–5. https://doi.org/10.1016/j.molp.2016.11.013.
Lv Y, Yang M, Hu D, Yang Z, Ma S, Li X, Xiong L. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression. Plant Physiol. 2017;173(2):1475–91. https://doi.org/10.1104/pp.16.01725.
Ly DNP, Iqbal S, Fosu-Nyarko J, Milroy S, Jones MGK. Multiplex CRISPR-Cas9 gene-editing can deliver potato cultivars with reduced browning and acrylamide. Plants (Basel). 2023;12(2):379. https://doi.org/10.3390/plants12020379.
Ma J, Chen J, Wang M, Ren Y, Wang S, Lei C, Cheng Z, Sodmergen. Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J Exp Bot. 2018;69(5):1051–64. https://doi.org/10.1093/jxb/erx458.
Macovei A, Sevilla NR, Cantos C, Jonson GB, Slamet-Loedin I, Čermák T, Voytas DF, Choi IR, Chadha-Mohanty P. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol J. 2018;16(11):1918–27. https://doi.org/10.1111/pbi.12927.
Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF. Plant gene editing through de novo induction of meristems. Nat Biotechnol. 2020;38(1):84–9. https://doi.org/10.1038/s41587-019-0337-2.
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR/Cas systems. Nat Rev Microbiol. 2015;13(11):722–36. https://doi.org/10.1038/nrmicro3569.
Mallapaty S. China’s approval of gene-edited crops energizes researchers. Nature. 2022;602(7898):559–60. https://doi.org/10.1038/d41586-022-00395-x.
Malnoy M, Viola R, Jung MH, Koo OJ, Kim S, Kim JS, Velasco R, Nagamangala KC. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci. 2016;7:1904. https://doi.org/10.3389/fpls.2016.01904.
Manghwar H, Li B, Ding X, Hussain A, Lindsey K, Zhang X, Jin S. CRISPR/Cas systems in genome editing: methodologies and tools for sgRNA design, off-target evaluation, and strategies to mitigate off-target effects. Adv Sci (Weinh). 2020;7(6):1902312. https://doi.org/10.1002/advs.201902312.
Mann A, Kumari J, Kumar R, Kumar P, Pradhan AK, Pental D, Bisht NC. Targeted editing of multiple homologues of GTR1 and GTR2 genes provides the ideal low-seed, high-leaf glucosinolate oilseed mustard with uncompromised defence and yield. Plant Biotechnol J. 2023. https://doi.org/10.1111/pbi.14121.
Metje-Sprink J, Menz J, Modrzejewski D, Sprink T. DNA-free genome editing: past, present and future. Front Plant Sci. 2019;9:1957. https://doi.org/10.3389/fpls.2018.01957.
Miki D, Zhang W, Zeng W, Feng Z, Zhu JK. CRISPR-Cas9 mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun. 2018;9(1):1967. https://doi.org/10.1038/s41467-018-04416-0.
Ming M, Ren Q, Pan C, He Y, Zhang Y, Liu S, Zhong Z, Wang J, Malzahn AA, Wu J, Zheng X, Zhang Y, Qi Y. CRISPR-Cas12b enables efficient plant genome engineering. Nat Plants. 2020;6(3):202–8. https://doi.org/10.1038/s41477-020-0614-6.
Mishra R, Zhao K. Genome editing technologies and their applications in crop improvement. Plant Biotechnol Rep. 2018;12:57–68. https://doi.org/10.1007/s11816-018-0472-0.
Modrzejewski D, Hartung F, Lehnert H, Sprink T, Kohl C, Keilwagen J, Wilhelm R. Which factors affect the occurrence of off-target effects caused by the use of CRISPR/Cas: a systematic review in plants. Front Plant Sci. 2020;11:574959. https://doi.org/10.3389/fpls.2020.574959.
Nagalakshmi U, Meier N, Liu JY, Voytas DF, Dinesh-Kumar SP. High-efficiency multiplex biallelic heritable editing in Arabidopsis using an RNA virus. Plant Physiol. 2022;189(3):1241–5. https://doi.org/10.1093/plphys/kiac159.
Nandy S, Pathak B, Zhao S, Srivastava V. Heat-shock-inducible CRISPR/Cas9 system generates heritable mutations in rice. Plant Direct. 2019;3(5):e00145. https://doi.org/10.1002/pld3.145.
Nawaz G, Han Y, Usman B, Liu F, Qin B, Li R. Knockout of OsPRP1, a gene encoding proline-rich protein, confers enhanced cold sensitivity in rice (Oryza sativa L.) at the seedling stage. 3 Biotech. 2019;9(7):254. https://doi.org/10.1007/s13205-019-1787-4.
Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep. 2017;7(1):482. https://doi.org/10.1038/s41598-017-00578-x.
Nguyen DV, Hoang TT, Le NT, Tran HT, Nguyen CX, Moon YH, Chu HH, Do PT. An efficient hairy root system for validation of plant transformation vector and CRISPR/Cas construct activities in cucumber (Cucumis sativus L.). Front Plant Sci. 2022;202212:770062. https://doi.org/10.3389/fpls.2021.770062.
Nieves-Cordones M, Mohamed S, Tanoi K, Kobayashi NI, Takagi K, Vernet A, Guiderdoni E, Périn C, Sentenac H, Véry AA. Production of low-Cs+ rice plants by inactivation of the K+ transporter OsHAK1 with the CRISPR/Cas system. Plant J. 2017;92(1):43–56. https://doi.org/10.1111/tpj.13632.
Nonaka S, Arai C, Takayama M, Matsukura C, Ezura H. Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci Rep. 2017;7(1):7057. https://doi.org/10.1038/s41598-017-06400-y.
Nuñez-Muñoz L, Vargas-Hernández B, Hinojosa-Moya J, Ruiz-Medrano R, Xoconostle-Cázares B. Plant drought tolerance provided through genome editing of the trehalase gene. Plant Signal Behav. 2021;16(4):1877005. https://doi.org/10.1080/15592324.2021.1877005.
O’Connell MR. Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR-Cas systems. J Mol Biol. 2019;431(1):66–87. https://doi.org/10.1016/j.jmb.2018.06.029.
Ogata T, Ishizaki T, Fujita M, Fujita Y. CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PLoS ONE. 2020;15(12):e0243376. https://doi.org/10.1371/journal.pone.0243376.
Okuzaki A, Ogawa T, Koizuka C, Kaneko K, Inaba M, Imamura J, Koizuka N. CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol Biochem. 2018;131:63–9. https://doi.org/10.1016/j.plaphy.2018.04.025.
Ortigosa A, Gimenez-Ibanez S, Leonhardt N, Solano R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol J. 2019;17(3):665–73. https://doi.org/10.1111/pbi.13006.
Osakabe Y, Watanabe T, Sugano SS, Ueta R, Ishihara R, Shinozaki K, Osakabe K. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci Rep. 2016;26(6):26685. https://doi.org/10.1038/srep26685.
Pan C, Li G, Malzahn AA, Cheng Y, Leyson B, Sretenovic S, Gurel F, Coleman GD, Qi Y. Boosting plant genome editing with a versatile CRISPR-Combo system. Nat Plants. 2022;8(5):513–25. https://doi.org/10.1038/s41477-022-01151-9.
Park JJ, Dempewolf E, Zhang W, Wang ZY. RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PLoS ONE. 2017;12(6):e0179410. https://doi.org/10.1371/journal.pone.0179410.
Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF, Doudna JA. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. 2020;369(6501):333–7. https://doi.org/10.1126/science.abb1400.
Peng A, Chen S, Lei T, Xu L, He Y, Wu L, Yao L, Zou X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J. 2017;15(12):1509–19. https://doi.org/10.1111/pbi.12733.
Pixley KV, Falck-Zepeda JB, Paarlberg RL, Phillips PWB, Slamet-Loedin IH, Dhugga KS, Campos H, Gutterson N. Genome-edited crops for improved food security of smallholder farmers. Nat Genet. 2022;54(4):364–7. https://doi.org/10.1038/s41588-022-01046-7.
Pyott DE, Sheehan E, Molnar A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol Plant Pathol. 2016;17(8):1276–88. https://doi.org/10.1111/mpp.12417.
Qin L, Li J, Wang Q, Xu Z, Sun L, Alariqi M, Manghwar H, Wang G, Li B, Ding X, Rui H, Huang H, Lu T, Lindsey K, Daniell H, Zhang X, Jin S. High-efficient and precise base editing of C•G to T•A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol J. 2020;18(1):45–56. https://doi.org/10.1111/pbi.13168.
Qiu Z, Kang S, He L, Zhao J, Zhang S, Hu J, Zeng D, Zhang G, Dong G, Gao Z, Ren D, Chen G, Guo L, Qian Q, Zhu L. The newly identified heat-stress sensitive albino 1 gene affects chloroplast development in rice. Plant Sci. 2018;267:168–79. https://doi.org/10.1016/j.plantsci.2017.11.015.
Rath D, Amlinger L, Rath A, Lundgren M. The CRISPR/Cas immune system: biology, mechanisms, and applications. Biochimie. 2015;117:119–28. https://doi.org/10.1016/j.biochi.2015.03.025.
Rathnasamy SA, Kambale R, Elangovan A, Mohanavel W, Shanmugavel P, Ramasamy G, Alagarsamy S, Marimuthu R, Rajagopalan VR, Manickam S, Ramanathan V, Muthurajan R, Vellingiri G. Altering stomatal density for manipulating transpiration and photosynthetic traits in rice through CRISPR/Cas9 mutagenesis. Curr Issues Mol Biol. 2023;45(5):3801–14. https://doi.org/10.3390/cimb45050245.
Reed KM, Bargmann BOR. Protoplast regeneration and its use in new plant breeding technologies. Front Genome Ed. 2021;3:734951. https://doi.org/10.3389/fgeed.2021.734951.
Ren B, Liu L, Li S, Kuang Y, Wang J, Zhang D, Zhou X, Lin H, Zhou H. Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing Ng and other atypical PAMs in rice. Mol Plant. 2019;12(7):1015–26. https://doi.org/10.1016/j.molp.2019.03.010.
Ren B, Yan F, Kuang Y, Li N, Zhang D, Zhou X, Lin H, Zhou H. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol Plant. 2018;11(4):623–6. https://doi.org/10.1016/j.molp.2018.01.005.
Roca Paixão JF, Gillet FX, Ribeiro TP, Bournaud C, Lourenço-Tessutti IT, Noriega DD, Melo BP, de Almeida-Engler J, Grossi-de-Sa MF. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a histone AcetylTransferase. Sci Rep. 2019;9(1):8080. https://doi.org/10.1038/s41598-019-44571-y.
Saber Sichani A, Ranjbar M, Baneshi M, Torabi Zadeh F, Fallahi J. A review on advanced CRISPR-based genome-editing tools: base editing and prime editing. Mol Biotechnol. 2023;65(6):849–60. https://doi.org/10.1007/s12033-022-00639-1.
Sánchez-León S, Gil-Humanes J, Ozuna CV, Giménez MJ, Sousa C, Voytas DF, Barro F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J. 2018;16(4):902–10. https://doi.org/10.1111/pbi.12837.
Santillán Martínez MI, Bracuto V, Koseoglou E, Appiano M, Jacobsen E, Visser RGF, Wolters AA, Bai Y. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 2020;20(1):284. https://doi.org/10.1186/s12870-020-02497-y.
Santosh Kumar VV, Verma RK, Yadav SK, Yadav P, Watts A, Rao MV, Chinnusamy V. CRISPR/Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol Mol Biol Plants. 2020;26(6):1099–110. https://doi.org/10.1007/s12298-020-00819-w.
Sashidhar N, Harloff HJ, Potgieter L, Jung C. Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol J. 2020;18(11):2241–50. https://doi.org/10.1111/pbi.13380.
Sauer NJ, Mozoruk J, Miller RB, Warburg ZJ, Walker KA, Beetham PR, Schöpke CR, Gocal GF. Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotechnol J. 2016;14(2):496–502. https://doi.org/10.1111/pbi.12496.
Schmidt C, Pacher M, Puchta H. Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J. 2019;98(4):577–89. https://doi.org/10.1111/tpj.14322.
Schwartz C, Lenderts B, Feigenbutz L, Barone P, Llaca V, Fengler K, Svitashev S. CRISPR-Cas9-mediated 75.5-Mb inversion in maize. Nat Plants. 2020;6(12):1427–31. https://doi.org/10.1038/s41477-020-00817-6.
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C. Targeted genome modification of crop plants using a CRISPR/Cas system. Nat Biotechnol. 2013;31(8):686–8. https://doi.org/10.1038/nbt.2650.
Sharma SK, Gupta OP, Pathaw N, Sharma D, Maibam A, Sharma P, Sanasam J, Karkute SG, Kumar S, Bhattacharjee B. CRISPR-Cas-Led revolution in diagnosis and management of emerging plant viruses: new avenues toward food and nutritional security. Front Nutr. 2021;8:751512. https://doi.org/10.3389/fnut.2021.751512.
Shen L, Wang C, Fu Y, Wang J, Liu Q, Zhang X, Yan C, Qian Q, Wang K. QTL editing confers opposing yield performance in different rice varieties. J Integr Plant Biol. 2018;60(2):89–93. https://doi.org/10.1111/jipb.12501.
Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H, Habben JE. ARGOS8 variants generated by CRISPR/Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15(2):207–16. https://doi.org/10.1111/pbi.12603.
Silva CJ, van den Abeele C, Ortega-Salazar I, Papin V, Adaskaveg JA, Wang D, Casteel CL, Seymour GB, Blanco-Ulate B. Host susceptibility factors render ripe tomato fruit vulnerable to fungal disease despite active immune responses. J Exp Bot. 2021;72(7):2696–709. https://doi.org/10.1093/jxb/eraa601.
Singh S, Chaudhary R, Deshmukh R, Tiwari S. Opportunities and challenges with CRISPR-Cas mediated homologous recombination based precise editing in plants and animals. Plant Mol Biol. 2023;111(1–2):1–20. https://doi.org/10.1007/s11103-022-01321-5.
Song X, Liu C, Wang N, Huang H, He S, Gong C, Wei Y. Delivery of CRISPR/Cas systems for cancer gene therapy and immunotherapy. Adv Drug Deliv Rev. 2021;168:158–80. https://doi.org/10.1016/j.addr.2020.04.010.
Sun SK, Xu X, Tang Z, Tang Z, Huang XY, Wirtz M, Hell R, Zhao FJ. A molecular switch in sulfur metabolism to reduce arsenic and enrich selenium in rice grain. Nat Commun. 2021;12(1):1392. https://doi.org/10.1038/s41467-021-21282-5.
Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015;169(2):931–45. https://doi.org/10.1104/pp.15.00793.
Taleei R, Nikjoo H. Biochemical DSB-repair model for mammalian cells in G1 and early S phases of the cell cycle. Mutat Res. 2013;756:206–12. https://doi.org/10.1016/j.mrgentox.2013.06.004.
Tamilselvan-Nattar-Amutha S, Hiekel S, Hartmann F, Lorenz J, Dabhi RV, Dreissig S, Hensel G, Kumlehn J, Heckmann S. Barley stripe mosaic virus-mediated somatic and heritable gene editing in barley (Hordeum vulgare L.). Front Plant Sci. 2023;14:1201446. https://doi.org/10.3389/fpls.2023.1201446.
Tan J, Zeng D, Zhao Y, Wang Y, Liu T, Li S, Xue Y, Luo Y, Xie X, Chen L, Liu YG, Zhu Q. PhieABEs: a PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol J. 2022;20(5):934–43. https://doi.org/10.1111/pbi.13774.
Tang XD, Gao F, Liu MJ, Fan QL, Chen DK, Ma WT. Methods for enhancing clustered regularly interspaced short palindromic repeats/Cas9-mediated homology-directed repair efficiency. Front Genet. 2019;10:551. https://doi.org/10.3389/fgene.2019.00551.
Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3:17018. https://doi.org/10.1038/nplants.2017.18.
Tang L, Mao B, Li Y, Lv Q, Zhang L, Chen C, He H, Wang W, Zeng X, Shao Y, Pan Y, Hu Y, Peng Y, Fu X, Li H, Xia S, Zhao B. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci Rep. 2017;7(1):14438. https://doi.org/10.1038/s41598-017-14832-9.
Tang X, Ren Q, Yang L, Bao Y, Zhong Z, He Y, Liu S, Qi C, Liu B, Wang Y, Sretenovic S, Zhang Y, Zheng X, Zhang T, Qi Y, Zhang Y. Single transcript unit CRISPR 20 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol J. 2019;17(7):1431–45. https://doi.org/10.1111/pbi.13068.
Thomazella DPT, Seong K, Mackelprang R, Dahlbeck D, Geng Y, Gill US, Qi T, Pham J, Giuseppe P, Lee CY, Ortega A, Cho MJ, Hutton SF, Staskawicz B. Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc Natl Acad Sci USA. 2021. https://doi.org/10.1073/pnas.2026152118.
Tian S, Jiang L, Cui X, Zhang J, Guo S, Li M, Zhang H, Ren Y, Gong G, Zong M, Liu F, Chen Q, Xu Y. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018;37(9):1353–6. https://doi.org/10.1007/s00299-018-2299-0.
Tian Y, Shen R, Li Z, Yao Q, Zhang X, Zhong D, Tan X, Song M, Han H, Zhu JK, Lu Y. Efficient C-to-G editing in rice using an optimized base editor. Plant Biotechnol J. 2022;20(7):1238–40. https://doi.org/10.1111/pbi.13841.
Tran MT, Doan DTH, Kim J, Song YJ, Sung YW, Das S, Kim EJ, Son GH, Kim SH, Van Vu T, Kim JY. CRISPR/Cas9-based precise excision of SlHyPRP1 domain(s) to obtain salt stress-tolerant tomato. Plant Cell Rep. 2021;40(6):999–1011. https://doi.org/10.1007/s00299-020-02622-z.
Tripathi JN, Ntui VO, Shah T, Tripathi L. CRISPR/Cas9-mediated editing of DMR6 orthologue in banana (Musa spp.) confers enhanced resistance to bacterial disease. Plant Biotechnol J. 2021;19(7):1291–3. https://doi.org/10.1111/pbi.13614.
Usman B, Nawaz G, Zhao N, Liao S, Liu Y, Li R. Precise editing of the OsPYL9 gene by RNA-guided Cas9 nuclease confers enhanced drought tolerance and grain yield in rice (Oryza sativa L.) by regulating circadian rhythm and abiotic stress responsive proteins. Int J Mol Sci. 2020;21(21):7854. https://doi.org/10.3390/ijms21217854.
Veillet F, Perrot L, Guyon-Debast A, Kermarrec MP, Chauvin L, Chauvin JE, Gallois JL, Mazier M, Nogué F. Expanding the CRISPR toolbox in P. patens using SpCas9-NG variant and application for gene and base editing in Solanaceae crops. Int J Mol Sci. 2020;21(3):1024. https://doi.org/10.3390/ijms21031024.
Vlčko T, Ohnoutková L. Allelic variants of CRISPR/Cas9 induced mutation in an Inositol Trisphosphate 5/6 Kinase gene manifest different phenotypes in barley. Plants (Basel). 2020;9(2):195. https://doi.org/10.3390/plants9020195.
Wang Q, Alariqi M, Wang F, Li B, Ding X, Rui H, Li Y, Xu Z, Qin L, Sun L, Li J, Zou J, Lindsey K, Zhang X, Jin S. The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol J. 2020;18(12):2436–43. https://doi.org/10.1111/pbi.13417.
Wang L, Chen L, Li R, Zhao R, Yang M, Sheng J, Shen L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J Agric Food Chem. 2017;65(39):8674–82. https://doi.org/10.1021/acs.jafc.7b02745.
Wang FZ, Chen MX, Yu LJ, Xie LJ, Yuan LB, Qi H, Xiao M, Guo W, Chen Z, Yi K, Zhang J, Qiu R, Shu W, Xiao S, Chen QF. OsARM1, an R2R3 MYB transcription factor, is involved in regulation of the response to arsenic stress in rice. Front Plant Sci. 2017;8:1868. https://doi.org/10.3389/fpls.2017.01868.
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–51. https://doi.org/10.1038/nbt.2969.
Wang T, Deng Z, Zhang X, Wang H, Wang Y, Liu X, Liu S, Xu F, Li T, Fu D, Zhu B, Luo Y, Zhu H. Tomato DCL2b is required for the biosynthesis of 22-nt small RNAs, the resulting secondary siRNAs, and the host defense against ToMV. Hortic Res. 2018;5:62. https://doi.org/10.1038/s41438-018-0073-7.
Wang Z, Hardcastle TJ, Canto Pastor A, Yip WH, Tang S, Baulcombe DC. A novel DCL2-dependent miRNA pathway in tomato affects susceptibility to RNA viruses. Genes Dev. 2018;32(17–18):1155–60. https://doi.org/10.1101/gad.313601.118.
Wang J, He Z, Wang G, Zhang R, Duan J, Gao P, Lei X, Qiu H, Zhang C, Zhang Y, Yin H. Efficient targeted insertion of large DNA fragments without DNA donors. Nat Methods. 2022;19(3):331–40. https://doi.org/10.1038/s41592-022-01399-1.
Wang W, Pan Q, Tian B, He F, Chen Y, Bai G, Akhunova A, Trick HN, Akhunov E. Gene editing of the wheat homologs of TONNEAU1-recruiting motif encoding gene affects grain shape and weight in wheat. Plant J. 2019;100(2):251–64. https://doi.org/10.1111/tpj.14440.
Wang H, La Russa M, Qi LS. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem. 2016;85:227–64. https://doi.org/10.1146/annurev-biochem-060815-014607.
Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, Liu YG, Zhao K. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the erf transcription factor gene OsERF922. PLoS ONE. 2016. https://doi.org/10.1371/journal.pone.0154027.
Wang M, Xu Z, Gosavi G, Ren B, Cao Y, Kuang Y, Zhou C, Spetz C, Yan F, Zhou X, Zhou H. Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnol J. 2020;18(8):1645–7. https://doi.org/10.1111/pbi.13330.
Wang F, Xu Y, Li W, Chen Z, Wang J, Fan F, Tao Y, Jiang Y, Zhu QH, Yang J. Creating a novel herbicide-tolerance OsALS allele using CRISPR/Cas9-mediated gene editing. Crop J. 2021;9:305–12. https://doi.org/10.1016/j.cj.2020.06.001.
Wang S, Yang Y, Guo M, Zhong C, Yan C, Sun S. Targeted mutagenesis of amino acid transporter genes for rice quality improvement using the CRISPR/Cas9 system. Crop J. 2020;8:457–64. https://doi.org/10.1016/j.cj.2020.02.005.
Wei C, Wang C, Jia M, Guo HX, Luo PY, Wang MG, Zhu JK, Zhang H. Efficient generation of homozygous substitutions in rice in one generation utilizing an rABE8e base editor. J Integr Plant Biol. 2021;63(9):1595–9. https://doi.org/10.1111/jipb.13089.
Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJ, van der Oost J. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46(5):595–605. https://doi.org/10.1016/j.molcel.2012.03.018.
Wright DA, Li T, Yang B, Spalding MH. TALEN-mediated genome editing: prospects and perspectives. Biochem J. 2014;462(1):15–24. https://doi.org/10.1042/BJ20140295.
Wu F, Qiao X, Zhao Y, Zhang Z, Gao Y, Shi L, Du H, Wang L, Zhang YJ, Zhang Y, Liu L, Wang Q, Kong D. Targeted mutagenesis in Arabidopsis thaliana using CRISPR-Cas12b/C2c1. J Integr Plant Biol. 2020;62(11):1653–8. https://doi.org/10.1111/jipb.12944.
Wu J, Yan G, Duan Z, Wang Z, Kang C, Guo L, Liu K, Tu J, Shen J, Yi B, Fu T, Li X, Ma C, Dai C. Roles of the Brassica napus DELLA protein BnaA6.RGA, in modulating drought tolerance by interacting with the ABA signaling component BnaA10.ABF2. Front Plant Sci. 2020;11:577. https://doi.org/10.3389/fpls.2020.00577.
Xu Y, Lin Q, Li X, Wang F, Chen Z, Wang J, Li W, Fan F, Tao Y, Jiang Y, Wei X, Zhang R, Zhu QH, Bu Q, Yang J, Gao C. Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol J. 2021;19(1):11–3. https://doi.org/10.1111/pbi.13433.
Xu R, Yang Y, Qin R, Li H, Qiu C, Li L, Wei P, Yang J. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J Genet Genomics. 2016;43(8):529–32. https://doi.org/10.1016/j.jgg.2016.07.003.
Xu W, Yang Y, Yang B, Krueger CJ, Xiao Q, Zhao S, Zhang L, Kang G, Wang F, Yi H, Ren W, Li L, He X, Zhang C, Zhang B, Zhao J, Yang J. A design optimized prime editor with expanded scope and capability in plants. Nat Plants. 2022;8(1):45–52. https://doi.org/10.1038/s41477-021-01043-4.
Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H. Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant. 2018;11(4):631–4. https://doi.org/10.1016/j.molp.2018.02.008.
Yan D, Ren B, Liu L, Yan F, Li S, Wang G, Sun W, Zhou X, Zhou H. High-efficiency and multiplex adenine base editing in plants using new TadA variants. Mol Plant. 2021;14(5):722–31. https://doi.org/10.1016/j.molp.2021.02.007.
Yang T, Ali M, Lin L, Li P, He H, Zhu Q, Sun C, Wu N, Zhang X, Huang T, Li CB, Li C, Deng L. Recoloring tomato fruit by CRISPR/Cas9-mediated multiplex gene editing. Hortic Res. 2022;10(1):uhac214. https://doi.org/10.1093/hr/uhac214.
Yang SH, Kim E, Park H, Koo Y. Selection of the high efficient sgRNA for CRISPR-Cas9 to edit herbicide related genes, PDS, ALS, and EPSPS in tomato. Appl Biol Chem. 2022;65(1):13.
Yin X, Biswal AK, Dionora J, Perdigon KM, Balahadia CP, Mazumdar S, Chater C, Lin HC, Coe RA, Kretzschmar T, Gray JE, Quick PW, Bandyopadhyay A. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 2017;36(5):745–57. https://doi.org/10.1007/s00299-017-2118-z.
Yin Y, Qin K, Song X, Zhang Q, Zhou Y, Xia X, Yu J. BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physiol. 2018;59(11):2239–54. https://doi.org/10.1093/pcp/pcy146.
Young JK, Gasior SL, Jones S, Wang L, Navarro P, Vickroy B, Barrangou R. The repurposing of type I-E CRISPR-Cascade for gene activation in plants. Commun Biol. 2019;18(2):383. https://doi.org/10.1038/s42003-019-0637-6.
Yu Y, Leete TC, Born DA, Young L, Barrera LA, Lee SJ, Rees HA, Ciaramella G, Gaudelli NM. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat Commun. 2020;11(1):2052. https://doi.org/10.1038/s41467-020-15887-5.
Yu QH, Wang B, Li N, Tang Y, Yang S, Yang T, Xu J, Guo C, Yan P, Wang Q, Asmutola P. CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci Rep. 2017;7(1):11874. https://doi.org/10.1038/s41598-017-12262-1.
Yue E, Cao H, Liu B. OsmiR535, a potential genetic editing target for drought and salinity stress tolerance in Oryza sativa. Plants (Basel). 2020;9(10):1337. https://doi.org/10.3390/plants9101337.
Yuste-Lisbona FJ, Fernández-Lozano A, Pineda B, Bretones S, Ortíz-Atienza A, García-Sogo B, Müller NA, Angosto T, Capel J, Moreno V, Jiménez-Gómez JM, Lozano R. ENO regulates tomato fruit size through the floral meristem development network. Proc Natl Acad Sci USA. 2020;117(14):8187–95. https://doi.org/10.1073/pnas.1913688117.
Yuyu C, Aike Z, Pao X, Xiaoxia W, Yongrun C, Beifang W, Yue Z, Liaqat S, Shihua C, Liyong C, Yingxin Z. Effects of GS3 and GL3.1 for grain size editing by CRISPR/Cas9 in rice. Rice Sci. 2020;27(5):405–13.
Zeng Y, Wen J, Zhao W, Wang Q, Huang W. Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR/Cas9 system. Front Plant Sci. 2020;10:1663. https://doi.org/10.3389/fpls.2019.01663.
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–71. https://doi.org/10.1016/j.cell.2015.09.038.
Zhang Y, Bai Y, Wu G, Zou S, Chen Y, Gao C, Tang D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017;91(4):714–24. https://doi.org/10.1111/tpj.13599.
Zhang A, Liu Y, Wang F, Li T, Chen Z, Kong D, Bi J, Zhang F, Luo X, Wang J, Tang J, Yu X, Liu G, Luo L. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol Breed. 2019;39:47. https://doi.org/10.1007/s11032-019-0954-y.
Zhang Y, Qi Y. CRISPR enables directed evolution in plants. Genome Biol. 2019;20(1):83. https://doi.org/10.1186/s13059-019-1693-4.
Zhao Y, Zhang C, Liu W, Gao W, Liu C, Song G, Li WX, Mao L, Chen B, Xu Y, Li X, Xie C. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci Rep. 2016;6:23890. https://doi.org/10.1038/srep23890.
Zhou J, Peng Z, Long J, Sosso D, Liu B, Eom JS, Huang S, Liu S, Vera Cruz C, Frommer WB, White FF, Yang B. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015;82(4):632–43. https://doi.org/10.1111/tpj.12838.
Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35(5):438–40. https://doi.org/10.1038/nbt.3811.
Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP. De novo domestication of wild tomato using genome editing. Nat Biotechnol. 2018. https://doi.org/10.1038/nbt.4272.s.
Acknowledgements
The authors express their gratitude to the National Agri-Food Biotechnology Institute (NABI), Department of Biotechnology (DBT) for research facilities and support. Thankful to DBT for a junior research fellowship to SS and Council of Scientific and Industrial Research (CSIR) for a junior research fellowship to RC. The present work was also supported by the Biotechnology Industry Research Assistance Council (BIRAC) for a banana biofortification project grant to ST. SS and RC are thankful to the Regional Centre for Biotechnology for PhD registration. The authors would like to acknowledge the DBT-eLibrary Consortium (DelCON) for providing access to online journals.
Funding
This work was supported by the Core Grant of National Agri-Food Biotechnology Institute [IN].
Author information
Authors and Affiliations
Contributions
ST conceived and designed the idea. SS and RC performed the literature survey. SS, RC, VL and ST wrote the manuscript. ST contributed to the editing of the manuscript. All authors have read and agreed to the present version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Corresponding Editor: Kutubuddin Ali Molla; Reviewers: Praveen Awasthi, Muntazir Mushtaq
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, S., Chaudhary, R., Lokya, V. et al. Genome editing based trait improvement in crops: current perspective, challenges and opportunities. Nucleus (2024). https://doi.org/10.1007/s13237-024-00472-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13237-024-00472-8