Abstract
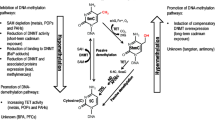
Humans can absorb volatile organic compounds (VOCs), such as ethylbenzene (EB), via inhalation resulting in various effects such as hepatotoxicity and even carcinogenicity. Because occupational EB exposure occurs frequently in the synthetic rubber and plastic industry, its toxicity has been previously reported by various in vitro as well as in vivo animal studies. EB has also been classified as a group 2B possible carcinogen by the IARC. However, these studies investigated the toxicity of EB used a much higher dose than expected occupational exposure. Thus the results are likely to be far away from to be the basis for the validation of the toxicity of the EB exposure. Because the previous studies examine only phenotypes, it is still unclear what and how EB affects physiological pathways in the human body. Using microarray platform, we investigated the gene expression profiles and genomic methylation patterns of 66 human blood samples from employees of local plants that use EB. The affected genes were functionally analyzed using the DAVID Tool and IPA. The methylation array demonstrated that, compared to the control group, a total of 1446 genes were hypermethylated and 60 genes were hypomethylated. Among the 378 differentially expressed genes, 12 had proper epigenetic correlation with the altered methylation patterns. Our study demonstrated how EB exposure affects the human genome by investigating the gene expression profiles and DNA methylation profiles of blood cells. We expect that our results will help establish the genomic basis of the effect of EB exposure on humans.
Similar content being viewed by others
References
IARC. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans., Vol. 77 227-266 (IARC Press, 2000).
NTP. NTP Toxicology and Carcinogenesis Studies of Ethylbenzene (CAS No. 100-41-4) in F344/N Rats and B6C3F1 Mice (Inhalation Studies). Natl. Toxicol. Program Tech. Rep. Ser. 466, 1–231 (1999).
Mutti, A. et al. Brain dopamine as a target for solvent toxicity: effects of some monocyclic aromatic hydrocarbons. Toxicology 49, 77–82 (1988).
Andersson, K. et al. Production of discrete changes in dopamine and noradrenaline levels and turnover in various parts of the rat brain following exposure to xylene, ortho-, meta-, and para-xylene, and ethylbenzene. Toxicology and Applied Pharmacology 60, 535–548 (1981).
Toftgård, R. & Nilsen, O.G. Effects of xylene and xylene isomers on cytochrome P-450 and in vitro enzymatic activities in rat liver, kidney and lung. Toxicology 23, 197–212 (1982).
Lee, S.Y. et al. Development of rheumatoid arthritis specific HLA-DRB1 genotyping microarray. BioChip J. 8, 187–198 (2014).
Son, G.W., Kim, G.-D., Yang, H., Park, H.R. & Park, Y.S. Alteration of gene expression profile by melatonin in endothelial cells. BioChip J. 8, 91–101 (2014).
Lee, D.-H. et al. Rapid hemagglutinin subtyping of novel avian-origin influenza A (H7N9) virus using a diagnostic microarray. BioChip J. 8, 55–59 (2014).
An, Y.R. & Hwang, S.Y. Toxicology study with microRNA. Mol. Cell. Toxicol. 10, 127–134 (2014).
Chai, J.C., Park, S., Seo, H., Cho, S.Y. & Lee, Y.S. Identification of cancer-specific biomarkers by using microarray gene expression profiling. BioChip J. 7, 57–62 (2013).
Song, M.-K., Song, M., Choi, H.-S., Park, Y.-K. & Ryu, J.C. Discovery of a characteristic molecular signature by microarray analysis of whole-blood gene expression in workers exposed to volatile organic compounds. BioChip J. 7, 112–135 (2013).
Choi, H.-S., Song, M.-K., Lee, E. & Ryu, J.-C. The toxicogenomic study on Persistent Organic Pollutants (POPs) in human hepatoma cell line. BioChip J. 7, 17–28 (2013).
Yang, H. et al. An integrated analysis of microRNA and mRNA expression in salvianolic acid B-treated human umbilical vein endothelial cells. Mol. Cell. Toxicol. 9, 1–7 (2013).
Huang, D.W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2008).
Huang, D.W., Sherman, B.T. & Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research 37, 1–13 (2009).
Sarma, S.N., Kim, Y.-J. & Ryu, J.-C. Differential gene expression profiling in human promyelocytic leukemia cells treated with benzene and ethylbenzene. Mol. Cell. Toxicol. 4, 267–277 (2008).
Lewis, B.P., Burge, C.B. & Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
An, Y.R. et al. Identification of genetic/epigenetic biomarkers for supporting decision of VOCs exposure. BioChip J. 7, 1–5 (2013).
Tsai, M.-C., Spitale, R.C. & Chang, H.Y. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Research 71, 3–7 (2011).
Backes, W., Sequeira, D., Cawley, G. & Eyer, C. Relationship between hydrocarbon structure and induction of P450: effects on protein levels and enzyme activities. Xenobiotica 23, 1353–1366 (1993).
Bergeron, R.M. et al. Changes in the expression of cytochrome P450s 2B1, 2B2, 2E1, and 2C11 in response to daily aromatic hydrocarbon treatment. Toxicology and Applied Pharmacology 157, 1–8 (1999).
Fishbein, L. An overview of environmental and toxicological aspects of aromatic hydrocarbons IV. Ethylbenzene. Science of the Total Environment 44, 269–287 (1985).
Midorikawa, K. et al. Metabolic activation of carcinogenic ethylbenzene leads to oxidative DNA damage. Chemico-biological Interactions 150, 271–281 (2004).
Serron, S.C., Dwivedi, N. & Backes, W.L. Ethylbenzene induces microsomal oxygen free radical generation: antibody-directed characterization of the responsible cytochrome P450 enzymes. Toxicology and Applied Pharmacology 164, 305–311 (2000).
Sams, C., Loizou, G.D., Cocker, J. & Lennard, M.S. Metabolism of ethylbenzene by human liver microsomes and recombinant human cytochrome P450s (CYP). Toxicology Letters 147, 253–260 (2004).
Chang, F.-K., Mao, I.-F., Chen, M.-L. & Cheng, S.-F. Urinary 8-hydroxydeoxyguanosine as a biomarker of oxidative DNA damage in workers exposed to ethylbenzene. Annals of Occupational Hygiene, mer010 (2011).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, G.W., Hong, J.Y., Yu, SY. et al. Integrative analyses of differential gene expression and DNA methylation of ethylbenzene-exposed workers. BioChip J 9, 259–267 (2015). https://doi.org/10.1007/s13206-015-9310-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-015-9310-4