Abstract
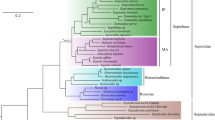
The molluscan clade Decapodiformes (Cephalopoda) comprises a diverse and enigmatic assemblage including inshore and offshore squids, bobtails, cuttlefishes, and the ram’s horn squid (Spirulida: Spirula spirula). The latter species is of particular interest to paleontologists because it is the only living cephalopod with an internal chambered, spiral-shaped, calcareous shell resembling those seen in some fossil cephalopod taxa. Spirulida has been difficult to place phylogenetically, in part because it shares different features with sepiolids, sepiids, and oegopsids, creating conflict in morphological analyses. Unlike morphological assumptions of a close relationship with sepiids, previous molecular studies have found support for Spirulida as a close relative of Bathyteuthida and Oegopsida. Identifying the correct phylogenetic placement of Spirulida could allow alternative hypotheses of phragmocone evolution, e.g., retention of an ancestral phragmocone in Spirulida and Sepiida vs. independent reacquisition of the phragmocone in these taxa, to be evaluated. In the present study, we combined new, high-quality transcriptome data for a specimen of Spirula spirula with additional new and previously published transcriptome data for decapodiform cephalopods. Phylogenetic analyses of several matrices yielded trees in which Spirula spirula was recovered as the sister group of the oceanic open-eyed squids (Oegopsida). This close relationship of Spirula to oceanic squids, rather than to nearshore, demersal/benthic decapodiform lineages, provides further support for an “onshore to offshore” model of decapodiform evolution.

Similar content being viewed by others
Data Availability
The datasets generated during and analyzed in this study, custom scripts used in the phylogenomics pipeline, and all trees are available in the Mendeley Data repository (http://dx.doi.org/10.17632/sk9jvz98vw.2).
References
Allcock, A. L., Lindgren, A., & Strugnell, J. M. (2014). The contribution of molecular data to our understanding of cephalopod evolution and systematics: A review. Journal of Natural History, 49(21–24), 1373–1421.
Anderson, F. E., & Lindgren, A. R. (2021). Phylogenomic analyses recover a clade of large-bodied decapodiform cephalopods. Molecular Phylogenetics and Evolution, 156, 107038. https://doi.org/10.1016/j.ympev.2020.107038
Ané, C., Larget, B., Baum, D. A., Smith, S. D., & Rokas, A. (2006). Bayesian estimation of concordance among gene trees. Molecular Biology and Evolution, 24(2), 412–426. https://doi.org/10.1093/molbev/msl170
Berry, S. S. (1911a). Preliminary notices of some new Pacific cephalopods. Proceedings of the United States National Museum, 4, 589–592.
Berry, S. S. (1911b). Note on a new Abraliopsis from Japan (A. scintillans n. sp.). Nautilus, 25, 93–94.
Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15), 2114–2120. https://doi.org/10.1093/BIOINFORMATICS/BTU170
Bushnell, B. (2015). BBMap short-read aligner, and other bioinformatics tools. https://sourceforge.net/projects/bbmap/
Carlini, D. B., & Graves, J. E. (1999). Phylogenetic analysis of cytochrome c oxidase I sequences to determine higher-level relationships within the coleoid cephalopods. Bulletin of Marine Science, 64, 57–76.
Chernomor, O., Von Haeseler, A., & Minh, B. Q. (2016). Terrace aware data structure for phylogenomic Inference from supermatrices. Systematic Biology, 65(6), 997–1008. https://doi.org/10.1093/SYSBIO/SYW037
Chikhi, R., & Medvedev, P. (2014). Informed and automated k-mer size selection for genome assembly. Bioinformatics, 30(1), 31–37. https://doi.org/10.1093/BIOINFORMATICS/BTT310
Clarke, M. R. (1969). Cephalopoda collected on the SOND Cruise. Journal of the Marine Biological Association of the United Kingdom, 49(4), 961–976. https://doi.org/10.1017/S0025315400038042
Cuvier, G. (1797). Tableau élémentaire de lhistoire naturelle des animaux. Baudouin, Paris, 710 pp.
Di Franco, A., Poujol, R., Baurain, D., & Philippe, H. (2019). Evaluating the usefulness of alignment filtering methods to reduce the impact of errors on evolutionary inferences. BMC Evolutionary Biology, 19(1), 1–17. https://doi.org/10.1186/S12862-019-1350-2
Donovan, D. T. (1977). Evolution of the dibranchiate cephalopoda. The biology of cephalopods (pp. 15–48). Academic Press.
Doyle, P., Donovan, D. T., & Nixon, M. (1994). Phylogeny and systematics of the Coleoidea. University of Kansas Paleontological Contributions, New Series, 5(1–15).
Dufayard, J.-F., Duret, L., Penel, S., Gouy, M., Rechenmann, F., & Perriere, G. (2005). Tree pattern matching in phylogenetic trees: Automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics, 21(11), 2596–2603. https://doi.org/10.1093/bioinformatics/bti325
Ebersberger, I., Strauss, S., & von Haeseler, A. (2009). HaMStR: Profile hidden markov model based search for orthologs in ESTs. BMC Evolutionary Biology, 9(1), 157. https://doi.org/10.1186/1471-2148-9-157
Emms, D. M., & Kelly, S. (2015). OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology, 16(1), 1–14. https://doi.org/10.1186/S13059-015-0721-2
Emms, D. M., & Kelly, S. (2019). OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biology, 20(1), 1–14. https://doi.org/10.1186/S13059-019-1832-Y
Fuchs, D., & Iba, Y. (2015). The gladiuses in coleoid cephalopods: Homology, parallelism, or convergence? Swiss Journal of Palaeontology, 134(2), 187–197. https://doi.org/10.1007/S13358-015-0100-3
Fuchs, D., Keupp, H., Trask, P., & Tanabe, K. (2012). Taxonomy, morphology and phylogeny of Late Cretaceous spirulid coleoids (Cephalopoda) from Greenland and Canada. Palaeontology, 55(2), 285–303.
Fuchs, D., Reitano, A., Insacco, G., & Iba, Y. (2016). The first coleoid cephalopods from the Upper Cenomanian of Sicily (Italy) and their implications for the systematic-phylogenetic position of the Palaeololiginidae (Teudopseina). Journal of Systematic Palaeontology, 15(6), 499–512. https://doi.org/10.1080/14772019.2016.1199055
Gadagkar, S. R., Rosenberg, M. S., & Kumar, S. (2005). Inferring species phylogenies from multiple genes: Concatenated sequence tree versus consensus gene tree. Journal of Experimental Zoology Part b: Molecular and Developmental Evolution, 304(1), 64–74.
Ganote, C. L., Sanders, S. A., Papudeshi, B. N., Blood, P. D., & Doak, T. G. (2017). A voice for bioinformatics. In Proceedings of the Practice and Experience in Advanced Research Computing 2017 on Sustainability, Success and Impact (p. 36). ACM.
Gould, S. J. (1970). Dollo on Dollo’s law: Irreversibility and the status of evolutionary laws. Journal of the History of Biology, 3(2), 189–212.
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology, 29(7), 644–652. https://doi.org/10.1038/nbt.1883
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., & Vinh, L. S. (2018). UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution, 35(2), 518–522. https://doi.org/10.1093/molbev/msx281
Hopkins, R. (2018). Evolution: Flip-flopping flower color defies Dollo’s Law. Current Biology, 28(23), R1337–R1339.
Hoyle, W. E. (1885). Narrative of the challenger expedition. Report on the scientific results of the voyage of HMS Challenger during the years 1873–1876. Narrative, 1(1), 269–274.
Joubin, L. (1898). Note sur une nouvelle famille de Céphalopodes. Annales des Sciences Naturelles, Série 8. Zoologie Et Biologie Animale, 6, 279–292.
Katoh, K., Kuma, K., Toh, H., & Miyata, T. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Research, 33(2), 511–518. https://doi.org/10.1093/nar/gki198
Kocot, K. M., Struck, T. H., Merkel, J., Waits, D. S., Todt, C., Brannock, P. M., et al. (2017). Phylogenomics of Lophotrochozoa with consideration of systematic error. Systematic Biology, 66(2), 256–282.
Kohlsdorf, T., & Wagner, G. P. (2006). Evidence for the reversibility of digit loss: A phylogenetic study of limb evolution in Bachia (Gymnophthalmoda: Squamata). Evolution, 60(9), 1896–1912. https://doi.org/10.1111/J.0014-3820.2006.TB00533.X
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., & Stamatakis, A. (2019). RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics, 35(21), 4453–4455. https://doi.org/10.1093/BIOINFORMATICS/BTZ305
Kück, P., & Meusemann, K. (2010). FASconCAT: Convenient handling of data matrices. Molecular Phylogenetics and Evolution, 56(3), 1115–1118. https://doi.org/10.1016/j.ympev.2010.04.024
Kück, P., & Struck, T. H. (2014). BaCoCa–A heuristic software tool for the parallel assessment of sequence biases in hundreds of gene and taxon partitions. Molecular Phylogenetics and Evolution, 70, 94–98. https://doi.org/10.1016/j.ympev.2013.09.011
Leach, W. E. (1817). Synopsis of the orders, families and genera of the class Cephalopoda. The Zoological Miscellany: being Descriptions of New, or Interesting Animals.
Lindgren, A. R., & Anderson, F. E. (2018). Assessing the utility of transcriptome data for inferring phylogenetic relationships among coleoid cephalopods. Molecular Phylogenetics and Evolution, 118, 330–342. https://doi.org/10.1016/j.ympev.2017.10.004
Lindgren, A. R., Giribet, G., & Nishiguchi, M. K. (2004). A combined approach to the phylogeny of Cephalopoda (Mollusca). Cladistics, 20(5), 454–486. https://doi.org/10.1111/J.1096-0031.2004.00032.X
Lindgren, A. R., Pankey, M. S., Hochberg, F. G., & Oakley, T. H. (2012). A multi-gene phylogeny of Cephalopoda supports convergent morphological evolution in association with multiple habitat shifts in the marine environment. BMC Evolutionary Biology, 12, 129. https://doi.org/10.1186/1471-2148-12-129
Lindsay, D. J., Hunt, J. C., McNeil, M., Beaman, R. J., & Vecchione, M. (2020). The first in situ observation of the ram’s horn squid Spirula spirula turns “common knowledge” upside down. Diversity, 12(12), 449. https://doi.org/10.3390/D12120449
MacManes, M. D. (2018). The Oyster River Protocol: A multi-assembler and kmer approach for de novo transcriptome assembly. PeerJ, 6(8), e5428. https://doi.org/10.7717/PEERJ.5428
Marshall, C. R., Raff, E. C., & Raff, R. A. (1994). Dollo’s law and the death and resurrection of genes. Proceedings of the National Academy of Sciences, 91(25), 12283–12287.
Miller, M., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Gateway Computing Environments Workshop (GCE), 2010 (pp. 1–8). IEEE.
Minh, B. Q., Dang, C. C., Vinh, L. S., & Lanfear, R. (2021). QMaker: Fast and accurate method to estimate empirical models of protein evolution. Systematic Biology, 70(5), 1046–1060. https://doi.org/10.1093/SYSBIO/SYAB010
Minh, B. Q., Hahn, M. W., & Lanfear, R. (2020). New methods to calculate concordance factors for phylogenomic datasets. Molecular Biology and Evolution, 37(9), 2727–2733. https://academic.oup.com/mbe/advancearticle/doi/10.1093/molbev/msaa106/5828940
Naef, A. (1922). Die Fossilen Tintenfische. Jena.
Naef, A. (1923). Cephalopoda. Fauna e Flora del Folgo di Napoli. Monograph No. 35. Teil I, Band 1, Lfg. 2. English translation: A Mercado (1972). Israel Program for Scientific Translations Ltd.
Ohkouchi, N., Tsuda, R., Chikaraishi, Y., & Tanabe, K. (2012). A preliminary estimate of the trophic position of the deep-water ram’s horn squid Spirula spirula based on the nitrogen isotopic composition of amino acids. Marine Biology, 160(4), 773–779. https://doi.org/10.1007/S00227-012-2132-1
Oudot, M., Neige, P., Shir, I. Ben, Schmidt, A., Strugnell, J. M., Plasseraud, L., et al. (2020). The shell matrix and microstructure of the Ram’s Horn squid: Molecular and structural characterization. Journal of Structural Biology, 211(1). https://doi.org/10.1016/j.jsb.2020.107507
Pfeffer, G. (1912). Die Cephalopoden der Plankton-Expedition: Zugleich eine monographische Übersicht der Oegopsiden Cephalopoden (Vol. 1). Lipsius & Tischer.
Price, M. N., Dehal, P. S., & Arkin, A. P. (2010). FastTree 2–Approximately maximum-likelihood trees for large alignments. PLoS ONE, 5(3), e9490. https://doi.org/10.1371/journal.pone.0009490
Quoy, J. R., & Gaimard, J. P. (1832). Mollusques, Voyage de decouvertes de l’Astrolabe pendant les annees 1826–1827-1828-1829. Zoologie, 2, 1–320.
Robertson, G., Schein, J., Chiu, R., Corbett, R., Field, M., Jackman, S. D., et al. (2010). De novo assembly and analysis of RNA-seq data. Nature Methods, 7(11), 909–912. https://doi.org/10.1038/nmeth.1517
Schrempf, D., Lartillot, N., & Szöllősi, G. (2020). Scalable empirical mixture models that account for across-site compositional heterogeneity. Molecular Biology and Evolution, 37(12), 3616–3631. https://doi.org/10.1093/molbev/msaa145
Shen, X. X., Li, Y., Hittinger, C. T., Chen, X. xin, & Rokas, A. (2020). An investigation of irreproducibility in maximum likelihood phylogenetic inference. Nature Communications, 11(1). https://doi.org/10.1038/s41467-020-20005-6
Si Quang, L., Gascuel, O., & Lartillot, N. (2008). Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics, 24(20), 2317–2323. https://doi.org/10.1093/bioinformatics/btn445
Song, L., & Florea, L. (2015). Rcorrector: Efficient and accurate error correction for Illumina RNA-seq reads. GigaScience, 4(1), 48. https://doi.org/10.1186/S13742-015-0089-Y
Sowerby, G. B. I (1821–1834) The genera of recent and fossil shells, for the use of students, in conchology and geology. Published in, 42, 1–126.
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9), 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Struck, T. H. (2014). TreSpEx—Detection of misleading signal in phylogenetic reconstructions based on tree information. Evolutionary Bioinformatics, 10, 51. https://doi.org/10.4137/EBO.S14239
Strugnell, J. M., Hall, N. E., Vecchione, M., Fuchs, D., & Allcock, A. L. (2017). Whole mitochondrial genome of the ram’s horn squid shines light on the phylogenetic position of the monotypic order Spirulida (Haeckel, 1896). Molecular Phylogenetics and Evolution, 109, 296–301. https://doi.org/10.1016/j.ympev.2017.01.011
Strugnell, J., & Nishiguchi, M. K. (2007). Molecular phylogeny of coleoid cephalopods (Mollusca: Cephalopoda) inferred from three mitochondrial and six nuclear loci: A comparison of alignment, implied alignment and analysis methods. Journal of Molluscan Studies, 73(4), 399–410. https://doi.org/10.1093/mollus/eym038
Strugnell, J., Norman, M., Jackson, J., Drummond, A. J., & Cooper, A. (2005). Molecular phylogeny of coleoid cephalopods (Mollusca: Cephalopoda) using a multigene approach; the effect of data partitioning on resolving phylogenies in a Bayesian framework. Molecular Phylogenetics and Evolution, 37(2), 426–441. https://doi.org/10.1016/J.YMPEV.2005.03.020
Sutton, M., Perales-Raya, C., & Gilbert, I. (2016). A phylogeny of fossil and living neocoleoid cephalopods. Cladistics, 32(3), 297–307. https://doi.org/10.1111/cla.12131
Tange, O. (2011). Gnu parallel-the command-line power tool. The USENIX Magazine, 36(1), 42–47.
Tanner, A. R., Fuchs, D., Winkelmann, I. E., Gilbert, M. T. P., Pankey, M. S., Ribeiro, Â. M., et al. (2017). Molecular clocks indicate turnover and diversification of modern coleoid cephalopods during the Mesozoic Marine Revolution. Proceedings of the Royal Society of London B: Biological Sciences, 284(1850), 20162818. https://doi.org/10.1098/rspb.2016.2818
Uribe, J. E., & Zardoya, R. (2017). Revisiting the phylogeny of Cephalopoda using complete mitochondrial genomes. Journal of Molluscan Studies, 83(2), 133–144. https://doi.org/10.1093/mollus/eyw052
von Linnaeus, C. (1758). Systema naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis (Vol. Tom. 1–2). Holmiae.
Wang, H., Minh, B., Susko, E., & Roger, A. (2017). Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Systematic Biology, 67(2), 216–235. https://doi.org/10.1093/sysbio/syx068
Wang, H., Susko, E., & Roger, A. (2019). The relative importance of modeling site pattern heterogeneity versus partition-wise heterotachy in phylogenomic inference. Systematic Biology, 68(6), 1003–1019. https://doi.org/10.1093/SYSBIO/SYZ021
Warnke, K., & Keupp, H. (2005). Spirula—A window to the embryonic development of ammonoids? Morphological and molecular indications for a palaeontological hypothesis. Facies, 51(1), 60–65. https://doi.org/10.1007/S10347-005-0054-9
Warnke, K. M., Meyer, A., Ebner, B., & Lieb, B. (2011). Assessing divergence time of Spirulida and Sepiida (Cephalopoda) based on hemocyanin sequences. Molecular Phylogenetics and Evolution, 58(2), 390–394. https://doi.org/10.1016/j.ympev.2010.11.024
Wiens, J. J. (2011). Re‐evolution of lost mandibular teeth in frogs after more than 200 million years, and re‐evaluating Dollo’s law. Evolution: International Journal of Organic Evolution, 65(5), 1283–1296.
Young, R. E. (1972). The systematics and areal distribution of pelagic cephalopods from the seas off southern California. Smithsonian Contributions to Zoology, 97.
Young, R. E., Vecchione, M., & Donovan, D. T. (1998). The evolution of coleoid cephalopods and their present biodiversity and ecology. South African Journal of Marine Science-Suid-Afrikaanse Tydskrif Vir Seewetenskap, 20, 393–420.
Zhong, M., Hansen, B., Nesnidal, M., Golombek, A., Halanych, K. M., & Struck, T. H. (2011). Detecting the symplesiomorphy trap: A multigene phylogenetic analysis of terebelliform annelids. BMC Evolutionary Biology, 11(1), 369. https://doi.org/10.1186/1471-2148-11-369
Acknowledgements
We thank Heather Judkins for assistance with voucher specimens; Alistair Tanner, Rute da Fonseca, and Jan Strugnell for sharing data and transcriptome assemblies with us; and the Integrated Genomics Laboratory at Oregon Health and Sciences University, the National Center for Genome Analysis Support at Indiana University, the SIU Research Computing and Cyberinfrastructure team, and BigDawg High Performance Computing Cluster for assistance with our own assemblies.
Funding
This research is based in part upon work supported by the National Science Foundation under Grant No. ABI-1759906 to Indiana University and DEB-103651 to Frank E. Anderson. Additional support was provided to Annie R Lindgren by a Portland State University Faculty Enhancement Grant and an Oregon State Oceangoing Research Vessel grant.
Author information
Authors and Affiliations
Contributions
Conceptualization: Frank E. Anderson, Annie R. Lindgren; data curation: Frank E. Anderson; formal analysis: Frank E. Anderson, Annie R. Lindgren, Abigail M. Pratt; funding acquisition: Frank E. Anderson, Annie R. Lindgren; investigation: Frank E. Anderson, Annie R. Lindgren, Michael Vecchione; methodology: Frank E. Anderson; project administration: Frank E. Anderson, Annie R. Lindgren; resources: Frank E. Anderson, Annie R. Lindgren; software: Frank E. Anderson; supervision: Frank E. Anderson, Annie R. Lindgren; writing — original draft: Frank E. Anderson, Annie R. Lindgren, Abigail M. Pratt, Michael Vecchione; writing — review and editing: Frank E. Anderson, Annie R. Lindgren, Abigail M. Pratt, Michael Vecchione.
Corresponding authors
Ethics declarations
Ethics approval
No approval of research ethics committees was required to accomplish the goals of this study because experimental work was conducted with tissues from unregulated invertebrate species.
Competing interests
The authors declare no competing interests.
Disclaimer
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation, the National Center for Genome Analysis Support, or Indiana University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lindgren, A.R., Pratt, A., Vecchione, M. et al. Finding a home for the ram’s horn squid: phylogenomic analyses support Spirula spirula (Cephalopoda: Decapodiformes) as a close relative of Oegopsida. Org Divers Evol 23, 91–101 (2023). https://doi.org/10.1007/s13127-022-00583-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-022-00583-7