Abstract
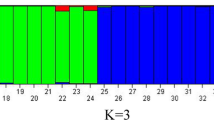
Genetic information of germplasm is the initial requirement for crop breeding programs. Rice is one of the oldest domesticated crop species endowed with rich genetic diversity which accounts for over 100,000 landraces and improved cultivars. The aim of the present study was to evaluate the genetic assessment of rice germplasm originating from India, the Philippines, China, and Malaysia through simple sequence repeat (SSR) markers. About 64 alleles were produced over 24 SSR primer amplifications over the whole genome of rice. The number of alleles ranged from 1 to 4 with an average of 2.67. Out of 64 amplified bands, 58 bands were polymorphic and 6 were monomorphic bands. Most of the primers showed high polymorphic information content (PIC). The PIC value ranged from 0.53 to 0.87. The cluster analysis indicates that the 73 varieties originating from India, the Philippines, China, and Malaysia were grouped separately and made two major clusters (seven groups). Among the two major clusters, one cluster had 18 genotypes which originated from the Phillipines, and China with 29% similarity with other varieties originating from India and Malaysia. Further, it was divided into two subgroups which had eight genotypes and the other had 10 genotypes with 41% similarity among themselves. All 10 genotypes were international varieties suitable for cultivation in medium land ecosystems. The second major cluster had 55 varieties including commercial rice varieties originating from India and Malaysia. The genotype ‘Swarna’ and ‘Manaswini’ had 76% similarity with each other and 69% similarity with the ‘Bhanja’ & ‘Ghanteswari’ which might be the genome association. The second major cluster had 55 genotypes divided into two minor groups. The first group had one genotype, i.e. ‘IR 63141-B-18-B’ with 34% similarity with the other 54 genotypes. The second minor group (54 genotypes) again was divided into two groups; one group had five genotypes with 51% similarity. Another group had 49 genotypes divided into two sub-minor groups. Based on this study, the larger range of similarity values using SSR markers provides greater confidence for the assessment of genetic relationships among the varieties. These genotypes are suitable for cultivation in upland ecosystems. The information obtained from the SSR profile helps to identify the variety diagnostic markers in 73 rice germplasm accessions. The intra- and inter-variation might be useful for breeders to improve the rice varieties through selective breeding and cross breeding programs.
Similar content being viewed by others
References
Aguirre C, Alvarado R, Hinrichsen P. 2005 Identificación de cultivares y líneas de mejoramiento de arroz de Chile mediante amplificación de fragmentos polimórficos (AFLP). Agric. Téc. 65: 356–369
Byerlee D. 1996. Knowledge-Intensive Crop Management Technologies: Concepts, Impacts, and Prospects in Asian Agriculture. International Rice Research Conference, Bangkok, Thailand, 3-5 June, 1996
Bligh HFJ, Blackhall NW, Edwards KJ, McClung AM. 1999. Using amplified length polymorphisms and simple sequence length polymorphisms to identify cultivars of brown and white milled rice. Crop Sci. 39: 1715–1721
Chuan-Guang, Gui-Quan Z. 2010. SSR analysis of genetic diversity and the temporal trends of major commercial inbred indica rice cultivars in south China in 1949–2005. Acta Agron. Sin. 36: 1843–1852
Choudhary G, Ranjitkumar N, Surapaneni M, Deborah DA, Vipparla A, Anuradha G, Siddiq EA, Vemireddy LR. 2013. Molecular genetic diversity of major Indian rice cultivars over decadal periods. PLoS ONE 8: e66197. doi: 10.1371/journal.pone.0066197
Cuevas-Perez FE, Guimaraes EP, Berrio LE, Gonzales DI. 1992. Genetic base of the irrigated rice in Latin America and the Caribbean. Crop Sci. 32: 1054–1059
Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13–15
Fuentes JL, Escobar F, Alvarez A, Gallego G, Duque MC, Ferrer M, Deus JE, Tohme J. 1999. Analyses of genetic diversity in Cuban rice varieties using isozyme, RAPD and AFLP markers. Euphytica 109: 107–115
Garris AJ, Tai TH, Coburn JR, Kresovich S, McCouch S. 2005. Genetic structure and diversity in Oryza sativa L. Genetics 169:1631–1638
Gupta PK, Varshney RK, Sharma PC, Ramesh B. 1999. Molecular markers and their applications in wheat breeding. Plant Breed. 118: 369–390
Guimaraes EP, Borrero J, Ospina-Rey Y. 1995. Genetic diversity of upland rice germplasm distributed in Latin America. Pesqui. Agropecu. Bras. 31: 187–194
Jain S, Jain RK, McCouch SR. 2004. Genetic analysis of Indian aromatic and quality rice (Oryza sativa L.) germplasm using panels of fluorescently-labeled microsatellite markers. Theor. Appl. Genet. 109: 965–977
Jayamani P, Negrao S, Martins M, Macas B, Oliveria MM. 2007. Genetic relatedness of Portuguese rice accessions from diverse origins as assessed by microsatellite markers. Crop Sci. 47: 879–884
Jeung JU, Hwang HG, Moon HP, Jena KK. 2005. Fingerprinting temperate japonica and tropical indica rice genotypes by comparative analysis of DNA markers. Euphytica 146: 239–251
Kshirsagar SS, Rabha M, Samal KC, Bastia DN, Rout GR. 2014. Identification of variety diagnostic molecular markerof high-yielding rice varieties. Proc. Natl. Acad. Sci., India, Sect. B. Biol. Sci. 84: 389–396
McCouch SR, Chen X, Panaud O, Temnykh S, Xu Y, Cho YG, Huang N, Ishii T, Blair M. 1997. Microsatellite mapping and applications of SSLP’s in rice genetics and breeding. Plant Molecular Biology, 35: 89–99
Meti N, Samal KC, Bastia DN, Rout GR. 2013. Genetic diversity analysis in aromatic rice genotypes using microsatellite based on simple sequence repeat markers. Afr. J. Biotechnol. 12: 4238–4250
Nagaraju J, Kathirve M, Ramesh RK, Siddiq EA, Hasnain E. 2002. Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties by using fluorescencebased ISSR-PCR and SSR markers. Agric. Sci. 99: 5836–5841
Ni J, Colowit PM, Mackill DJ. 2002. Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci. 42: 601–607
Pervaiz ZH, Rabbani MA, Pearce SR, Malik SA. 2009. Determination of genetic variability of Asian rice (Oryza sativa L.) varieties using microsatellite markers. Afr. J. Biotech. 8: 5641–5651
Powel W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A. 1996. Comparison of RFLP RAPD, AFLP and SSR markers for germplasm analysis. Mol. Breed, 2(3): 225–238
Rangel PHN, Guimaraes EP, Neves PCF. 1996. The genetic base of Brazilian irrigated rice (Oryza sativa L) cultivars. Pesqu. Agropecu. Bras. 31: 349–357
Ray S, Agarwa P, Arora R, Kapoor S, Tyagi AK. 2007. Expression profile of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (oryza sativa L. ssp. Indica). Mol. Gen. Genomics 278: 493–505
Ren F, Lu BR, Li S, Huang J, Zhu Y. 2003. A comparative study of genetic relationships among the AA-genome Oryza species using RAPD and SSR markers. Theor. Appl. Genet. 108: 113–120
Rohlf FJ. 2002. NTSYS pc Numerical taxonomy and multivariate system Ver. 2.1 Exeter Publ. Ltd., Setauket, New York
Sajib AM, Hossain MM, Mosnaz ATMZ, Hossain H, Islam MM, Ali MS, Prodhan SH. 2012. SSR marker-based molecular characterization and genetic diversity analysis of aromatic land reces of rice (Oryza sativa L.). Jour. BioSci. Biotech. 1: 107–116
Shishido R, Kikuchi M, Nomura K, Ikehashi H. 2006. Evaluation of genetic diversity of wild rice (Oryza rufipogon Griff.) in Myanmar using simple sequence repeats (SSRs). Genet. Res. Crop Evol. 53: 179–186
Singh RK, Sharma RK, Singh AK, Singh VP, Singh NK, Tiwari SP, Mohapatra T. 2004. Suitability of mapped sequence tagged microsatellite site markers for establishing distinctness, uniformity and stability in aromatic rice. Euphytica 135: 135–143
Sneath PHA, Sokal RR. 1973. Numerical taxonomy. The principles and practice of numerical classification WH Freeman, San Francisco, CA
Spada A, Mantegazza R, Biloni M, Caporali E, Sala F. 2004. Italian rice varieties: historical data, molecular markers and pedigrees to reveal their genetic relationships. Plant Breed. 123: 105–111
Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR. 2000. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 100: 697–712
Xu Y, Beachell H, McCouch SR. 2004. A marker-based approach to broadening the genetic base of rice in USA. Crop Sci. 44: 1947–1959
Yu SB, Xu WJ, Vijayakumar CHM, Ali J, Fu BY, Xu JL, Jiang YZ, Marghirang R, Domingo J, Aqino C, Virmani SS, Li ZK. 2003. Molecular diversity and multilocus organization of the parental lines used in the International Rice Molecular Breeding Program. Theo. Appl. Genet.108: 131–140
Zhu MY, Wang YY, Zhu YY, Lu BR. 2004. Genetic diversity of rice landraces from Yunnan revealed by SSR analysis and its significance for conservation. J. Huazhong Agri. Univ. 23: 187–191
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nayak, S., Rajpalsingh, J.K., Bastia, D.N. et al. Assessment of seventy-three rice germplasm by using simple sequence repeats markers. J. Crop Sci. Biotechnol. 17, 297–304 (2014). https://doi.org/10.1007/s12892-014-0074-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-014-0074-5