Abstract
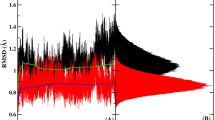
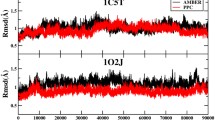
The effect of polarization in biomolecular force field is investigated by performing Molecular Dynamics (MD) simulation of HIV-protease by using two AMBER force fields, namely ff99 (non-polarizable) and ff02 (polarizable). The results of simulation show that the overall structural fluctuation of HIV-protease is reduced in the polarizable simulation. Comparison with the NMR order parameters with the calculated values shows that although some residues are less flexible in the ff02 simulation, the dynamics of two β-hairpins (flaps), the most flexible part of the protein, is relatively insensitive to the effect of polarization. The flap-active site distance, a measure of flap opening, is distinctly more in the non-polarizable simulation. The water count and radial distribution functions are investigated near a representative residue of three types — charged, polar and hydrophobic. Both water count and radial distribution function differ significantly near the charged residue (catalytic Asp25) between the force fields. However, the water movement is similar near the polar (Ser37) and hydrophobic (Ile85) residues. The preliminary results of this investigation show that polarization is likely to influence both global and specific local motions of protein and solvent.
Similar content being viewed by others
References
M Karplus and J A McCammon Nat. Struct. Biol. 9 646 (2002)
J Behler, R Martonak, D Donadio and M Parrinello Phys. Rev. Lett. 100 185501 (2008)
K Y Sanbonmatsu, S Joseph and C Tung PNAS 102 44 (2005)
B Chanda, O K Asamoah, R Blunck, B Roux and F Bezanilla Nature 436 852 (2005)
A Arkhipov, P L Freddolino, K Imada, K Namba and K Schulten Biophys. J. 91 4589 (2006)
Daan Frenkel and Berend Smit Understanding Molecular Simulation (Academic Press. 2nd ed.) (2001)
U H E Hansmann and Y Okamoto Curr. Opin. in Struct. Biol. 9 177 (1999)
F D Rienzo, R R Gabdoulline, M C Menziani, P G D Benedetti and R C Wade Biophys. J. 81 3090 (2001)
A D MacKerell, D Bashford, M Bellot, R L Dunbrack, J D Evanseck, M J Field, S Fischer, J Gao, H Guo, S Ha, D Joseph-MacCarthy, L Kuchnir, K Kuczera, F T K Lau, C Mattos, S Michnick, T Ngo, D T Nguyen, B Prodhom, W E I Reiher, B Roux, M Schlenkrich, J C Smith, R Stote, J Straub, M Watanabe, J Wiorkiewicz-Kuczera, D Yin and M Karplus J. Phys. Chem. A102 3586 (1998)
W D Cornell, P Cieplak, C I Bayly, I R Gould, K M Merz, D M Ferguson, D C Spellmeyer, T Fox, J W Caldwell and P A Kollman J. Am. Chem. Soc. 117 5179 (1995)
G A Kaminski, H A Stern, B J Berne, R A Friesner, Y X Cao, R B Murphy, R Zhou and T A Halgren J. Comput. Chem. 16 1515 (2002)
P Cieplak, J Caldwell and P A Kollman J. Comp. Chem. 22 1048 (2001)
P Ren and J W Ponder J. Phys. Chem. B107 5933 (2003)
B Kim, T Young, E Harder, R A Friesner and B J Berne J. Phys. Chem. B109 16529 (2005)
J Wang, P Cieplak and P Kollman J. Comp. Chem. 21 1049 (2000)
P Cieplak, J Caldwell and P Kollman J. Comp. Chem. 22 1048 (2001)
W R Scott and C A Schiffer Structure 8 1259 (2000)
V Hornak, A Okur, R C Rizzo and C Simmerling Proc. Natl. Acad. Sci. USA 103 915 (2006)
J R Collins, S K Burt and J W Erickson Nat. Str. Biol. 2 334 (1995)
A L Perryman, J H Lin and J A McCammon Protein Sci. 13 1108 (2004)
H Ode, S Neva, M Hata, W Sugiura and T Hoshino J. Am. Chem. Soc. 128 7887 (2006)
P Bandyopadhyay and B R Meher Chem. Biol. Drug. Des. 67 155 (2006)
W L Jorgensen, J Chandrasekhar, J Madura and M L Klein J. Chem. Phys. 79 926 (1983)
J W Caldwell and P A Kollman J. Phys. Chem. 99 6208 (1995)
D A Case, T A Darden, T E Cheathamlll, C L Simmerling, J Wang, R E Duke, R Luo, K M Merz, B Wang, D A Pearlman, M Crowley, S Brozell, V Tsui, H Gohlke, J Mongan, V Hornak, G Cui, P Beroza, C Schafmeister, J Caldwell, W Ross, P A Kollman AMBER 8 (San Francisco, CA: University of California) (2004)
S Spinelli, Q Z Liu, P M Alzari, P H Hirel and R J Poljak Biochimie 73 1391 (1991)
U Essmann, L Pereea, M L Berkowitz and T A Darden J. Chem. Phys. 103 8577 (1995)
H J C Berendsen, J P M Postma, W F Van Gunsteren, A Dinola and J R Haak J. Chem. Phys. 81 3684 (1984)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meher, B.R., Satish Kumar, M.V. & Bandyopadhyay, P. Molecular dynamics simulation of HIV-protease with polarizable and non-polarizable force fields. Indian J Phys 83, 81–90 (2009). https://doi.org/10.1007/s12648-009-0005-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12648-009-0005-3