Abstract
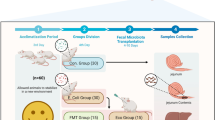
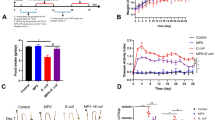
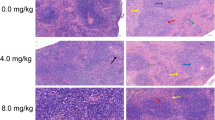
Giant pandas often suffered from gastrointestinal disease, especially the captive sub-adult one. Our study aims to investigate whether L. plantarum G83, a good panda-derived probiotic, can improve the intestinal barrier against the enterotoxigenic Escherichia coli K88 (E. coli K88) infection in giant panda microbiota-associated mice (GPAM). We treated SPF mice with antibiotics cocktail and transplanted the giant panda intestinal microbiota to set up a GPAM. Our results demonstrated that the microbiota of GPAM changed over time and was relatively stable in the short-term experiment (2–4 weeks). Whereafter, the GPAM pretreated with L. plantarum G83 for 15 days and infected with enterotoxigenic E. coli K88. The result indicated that the number of Bifidobacteria spp. increased in GPAM-G and GPAM-GE groups; the Lactobacillus spp. only increased in the GPAM-G group. Although the abundance of Enterobacteriaceae spp. only decreased in the GPAM-G group, the copy number of Escherichia coli in the GPAM-E group was significantly lower than that in the other groups. Meanwhile, the L. plantarum G83-induced alteration of microbiota could increase the mRNA expression of Claudin-1, Zo-1, and Occludin-1 in the GPAM-G group in the ileum; only Occludin-1 was increased in the GPAM-GE group. The sIgA in the ileum showed a positive response, also the result of body weight and histology in both the GPAM-G and GPAM-GE group. These results indicated that the L. plantarum G83 could improve the intestinal barrier to defense the enterotoxigenic E. coli K88 invasion.










Similar content being viewed by others
References
Ren L, Deng LH, Zhang RP, Wang CD, Li DS, Xi LX, Chen ZR, Yang R, Huang J, Zeng YR (2017) Relationship between drug resistance and the clustered, regularly interspaced, short, palindromic repeat-associated protein genes cas1 and cas2 in Shigella from giant panda dung. Medicine 96(7):e5922. https://doi.org/10.1097/MD.0000000000005922
Qiu X, Mainka SA (1993) Review of mortality of the giant panda (Ailuropoda melanoleuca). J Zoo Wildl Med 24(4):425–429 www.jstor.org/stable/20095301
Xue Z, Zhang W, Wang L, Hou R, Zhang M, Fei L, Zhang X, Huang H, Bridgewater LC, Jiang Y (2015) The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. Mbio 6(3):00022–00015. https://doi.org/10.1128/mBio.00022-15
Tingmei H, Tingting C, Zhijun Z, Chengdong W, Yongguo H, Yi W, Hemin Z, Desheng L, Caiwu L, Tingting Z, Yongjiu L, Anchun C, Guangneng P (2013) Research on diversity of intestinal flora in an adult giant panda based on the 16s rDNA-RFLP. Chin Vet Sci 42(11):1121–1127. https://doi.org/10.16656/j.issn.1673-4696.2012.11.019
Yongguo H, Lei J, Guo L, Caiwu L, Bei L, Wei L, Yahui Z, Zhengquan H, Yan H, Heming Z, Likou Z (2017) Gut microbiome of adult giant pandas based on high-throughput sequencing technology. Chin J of Appl Environ Biol 23(5):771–777. https://doi.org/10.3724/SP.J.1145.2016.05023
Zhang A, Wang H, Tian G, Zhang Y, Yang X, Xia Q, Tang J, Zou L (2009) Phenotypic and genotypic characterisation of antimicrobial resistance in faecal bacteria from 30 Giant pandas. Int J Antimicrob Ag 33(5):456–460. https://doi.org/10.1016/j.ijantimicag.2008.10.030
Zhang M, Zhang Z, Li Z, Hong M, Zhou X, Zhou S, Zhang J, Hull V, Huang J, Zhang H (2018) Giant panda foraging and movement patterns in response to bamboo shoot growth. Environ Sci Pollut Res Int 25(9):8636–8643. https://doi.org/10.1007/s11356-017-0919-9
Seres DS, Van Way CW (2015) Nutrition support for the critically ill. Curr Opin Gastroenterol 29(2):208–215. https://doi.org/10.1097/MOG.0b013e32835c9c83
Bastani P, Akbarzadeh F, Homayouni A, Javadi M, Khalili L (2016) Health benefits of probiotic consumption. In: Garg N, Abdel-Aziz S, Aeron A (eds) Microbes in food and health. Springer, Cham. https://doi.org/10.1007/978-3-319-25277-3_9
Shang Q, Shan X, Cai C, Hao J, Li G, Yu G (2016) Dietary fucoidan modulates the gut microbiota in mice by increasing the abundance of Lactobacillus and Ruminococcaceae. Food Funct 7(7):3224–3232. https://doi.org/10.1039/c6fo00309e
Balda MS, Matter K (2008) Tight junctions at a glance. J Cell Sci 121(22):3677–3682. https://doi.org/10.1242/jcs.023887
Feldman GJ, Mullin JM, Ryan MP (2005) Occludin: structure, function and regulation. Adv Drug Deliv Rev 57(6):883–917. https://doi.org/10.1016/j.addr.2005.01.009
Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M, Tsukita S (2006) ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126(4):741–754. https://doi.org/10.1016/j.cell.2006.06.043
Sánchez de Medina F, Romero-Calvo I, Mascaraque C, Martínez-Augustin O (2014) Intestinal inflammation and mucosal barrier function. Inflamm Bowel Dis 20(12):2394–2404. https://doi.org/10.1097/mib.0000000000000204
Sun H, Bi J, Lei Q, Wan X, Jiang T, Wu C, Wang X (2018) Partial enteral nutrition increases intestinal sIgA levels in mice undergoing parenteral nutrition in a dose-dependent manner. Int J Surg 49:74–79. https://doi.org/10.1016/j.ijsu.2017.12.011
Sánchez B, Delgado S, Blanco-Míguez A, Lourenço A, Gueimonde M, Margolles A (2017) Probiotics, gut microbiota, and their influence on host health and disease. Mol Nutr Food Res 61(1):1600240. https://doi.org/10.1002/mnfr.201600240
Wieërs G, Belkhir L, Enaud R, Leclercq S, Philippart de Foy J-M, Dequenne I, de Timary P, Cani PD (2020) How probiotics affect the microbiota. Front Cell Infect Microbiol 9:454. https://doi.org/10.3389/fcimb.2019.00454
Wang J, Ji H, Wang S, Liu H, Zhang W, Zhang D, Wang Y (2018) Probiotic Lactobacillus plantarum promotes intestinal barrier function by strengthening the epithelium and modulating gut microbiota. Front Microbiol 9:1953. https://doi.org/10.3389/fmicb.2018.01953
Wang L, Li L, Lv Y, Chen Q, Feng J, Zhao X (2018) Lactobacillus plantarum restores intestinal permeability disrupted by Salmonella infection in newly-hatched chicks. Sci Rep 8(1):2229. https://doi.org/10.1038/s41598-018-20752-z
Anderson RC, Cookson AL, McNabb WC, Park Z, McCann MJ, Kelly WJ, Roy NC (2010) Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol 10(1):316. https://doi.org/10.1186/1471-2180-10-316
Zihni C, Mills C, Matter K, Balda MS (2016) Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol 17(9):564–580. https://doi.org/10.1038/nrm.2016.80
Wang P, Li Y, Xiao H, Shi Y, Gw L, Sun J (2016) Isolation of Lactobacillus reuteri from Peyer’s patches and their effects on sIgA production and gut microbiota diversity. Mol Nutr Food Res 60(9):2020–2030. https://doi.org/10.1002/mnfr.201501065
Liu Q, Ni X, Wang Q, Peng Z, Niu L, Xie M, Lin Y, Zhou Y, Sun H, Pan K (2019) Investigation of lactic acid bacteria isolated from giant panda feces for potential probiotics in vitro. Probiotics Antimicrob Proteins 11(1):85–91. https://doi.org/10.1007/s12602-017-9381-8
Liu Q, Ni X, Wang Q, Peng Z, Niu L, Wang H, Zhou Y, Sun H, Pan K, Jing B (2017) Lactobacillus plantarum BSGP201683 isolated from giant panda feces attenuated inflammation and improved gut microflora in mice challenged with enterotoxigenic Escherichia coli. Front Microbiol 8:1885. https://doi.org/10.3389/fmicb.2017.01885
Zeng B, Li G, Yuan J, Li W, Tang H, Wei H (2013) Effects of age and strain on the microbiota colonization in an infant human flora-associated mouse model. Curr Microbiol 67(3):313–321. https://doi.org/10.1007/s00284-013-0360-3
Dal Bello F, Walter J, Hertel C, Hammes WP (2001) In vitro study of prebiotic properties of Levan-type exopolysaccharides from lactobacilli and non-digestible carbohydrates using denaturing gradient gel electrophoresis. Syst Appl Microbiol 24(2):232–237. https://doi.org/10.1078/0723-2020-00033
Qing X, Zeng D, Wang H, Ni X, Liu L, Lai J, Khalique A, Pan K, Jing B (2017) Preventing subclinical necrotic enteritis through Lactobacillus johnsonii BS15 by ameliorating lipid metabolism and intestinal microflora in broiler chickens. AMB Express 7(1):139. https://doi.org/10.1186/s13568-017-0439-5
Brandt LJ, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, Park T, Stollman N, Rohlke F, Surawicz C (2012) Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol 107(7):1079–1087. https://doi.org/10.1038/ajg.2012.60
Zhang X, Zeng B, Liu Z, Liao Z, Li W, Wei H, Fang X (2014) Comparative diversity analysis of gut microbiota in two different human flora-associated mouse strains. Curr Microbiol 69(3):365–373. https://doi.org/10.1007/s00284-014-0592-x
Becattini S, Taur Y, Pamer EG (2016) Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 22(6):458–478. https://doi.org/10.1016/j.molmed.2016.04.003
Buffie CG, Pamer EG (2013) Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13(11):790–801. https://doi.org/10.1038/nri3535
Nagao-Kitamoto H, Kitamoto S, Kuffa P, Kamada N (2016) Pathogenic role of the gut microbiota in gastrointestinal diseases. Intest Res 14(2):127–138. https://doi.org/10.5217/ir.2016.14.2.127
Ghoshal UC, Ghoshal U (2017) Small intestinal bacterial overgrowth and other intestinal disorders. Gastroenterol Clin N Am 46(1):103–120. https://doi.org/10.1016/j.gtc.2016.09.008
Blanchi J, Goret J, Mégraud F (2016) Clostridium difficile infection: a model for disruption of the gut microbiota equilibrium. Dig Dis 34(3):217–220. https://doi.org/10.1159/000443355
Bien J, Palagani V, Bozko P (2013) The intestinal microbiota dysbiosis and Clostridium difficile infection: is there a relationship with inflammatory bowel disease? Ther Adv Gastroenterol 6(1):53–68. https://doi.org/10.1177/1756283X12454590
Agus A, Denizot J, Thévenot J, Martinez-Medina M, Massier S, Sauvanet P, Bernalier-Donadille A, Denis S, Hofman P, Bonnet R (2016) Western diet induces a shift in microbiota composition enhancing susceptibility to adherent-invasive E. coli infection and intestinal inflammation. Sci Rep 6(19032):1–8. https://doi.org/10.1038/srep19032
Guo W, Mishra S, Zhao J, Tang J, Zeng B, Kong F, Ning R, Li M, Zhang H, Zeng Y (2018) Metagenomic study suggests that the gut microbiota of the giant panda (Ailuropoda melanoleuca) may not be specialized for fiber fermentation. Front Microbiol 9:229. https://doi.org/10.3389/fmicb.2018.00229
Guo L, Long M, Huang Y, Wu G, Deng W, Yang X, Li B, Meng Y, Cheng L, Fan L (2015) Antimicrobial and disinfectant resistance of Escherichia coli isolated from giant pandas. J Appl Microbiol 119(1):55–64. https://doi.org/10.1111/jam.12820
Candela M, Perna F, Carnevali P, Vitali B, Ciati R, Gionchetti P, Rizzello F, Campieri M, Brigidi P (2008) Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol 125(3):286–292. https://doi.org/10.1016/j.ijfoodmicro.2008.04.012
Collado MC, Grześkowiak Ł, Salminen S (2007) Probiotic strains and their combination inhibit in vitro adhesion of pathogens to pig intestinal mucosa. Curr Microbiol 55(3):260–265. https://doi.org/10.1007/s00284-007-0144-8
Suzuki C, Kimoto-Nira H, Kobayashi M, Nomura M, Sasaki K, Mizumachi K (2008) Immunomodulatory and cytotoxic effects of various Lactococcus strains on the murine macrophage cell line J774.1. Int J Food Microbiol 123(1): 159–165. https://doi.org/10.1016/j.ijfoodmicro.2007.12.022
Kimoto H, Mizumachi K, Okamoto T, J-i K (2013) New Lactococcus strain with immunomodulatory activity: enhancement of Th1-type immune response. Microbiol Immunol 48(2):75–82. https://doi.org/10.1111/j.1348-0421.2004.tb03490.x
Zhang L, Xu YQ, Liu HY, Lai T, Ma JL, Wang JF, Zhu YH (2010) Evaluation of Lactobacillus rhamnosus GG using an Escherichia coli K88 model of piglet diarrhoea: effects on diarrhoea incidence, faecal microflora and immune responses. Vet Microbiol 141(1–2):142–148. https://doi.org/10.1016/j.vetmic.2009.09.003
Liu Q, Ni X, Wang Q, Peng Z, Niu L, Xie M, Lin Y, Zhou Y, Sun H, Pan K (2018) Investigation of lactic acid bacteria isolated from giant panda feces for potential probiotics In Vitro. Probiotics Antimicrob Proteins 11(1): 85–91. https://doi.org/10.1007/s12602-017-9381-8
Yang F, Wang A, Zeng X, Hou C, Liu H, Qiao S (2015) Lactobacillus reuteri I5007 modulates tight junction protein expression in IPEC-J2 cells with LPS stimulation and in newborn piglets under normal conditions. BMC Microbiol 15(1):32. https://doi.org/10.1186/s12866-015-0372-1
Noth R, Lange-Grumfeld J, Stüber E, Kruse ML, Ellrichmann M, Häsler R, Hampe J, Bewig B, Rosenstiel P, Schreiber S (2011) Increased intestinal permeability and tight junction disruption by altered expression and localization of occludin in a murine graft versus host disease model. BMC Gastroenterol 11(1):109. https://doi.org/10.1186/1471-230X-11-109
Barreau F, Hugot J (2014) Intestinal barrier dysfunction triggered by invasive bacteria. Curr Opin Microbiol 17:91–98. https://doi.org/10.1016/j.mib.2013.12.003
Cording J, Berg J, Käding N, Bellmann C, Tscheik C, Westphal JK, Milatz S, Günzel D, Wolburg H, Piontek J (2013) In tight junctions, claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization. J Cell Sci 126(2):554–564. https://doi.org/10.1242/jcs.114306
Wesemann DR, Portuguese AJ, Meyers RM, Gallagher MP, Cluff-Jones K, Magee JM, Panchakshari RA, Rodig SJ, Kepler TB, Alt FW (2013) Microbial colonization influences early B-lineage development in the gut lamina propria. Nature 501(7465):112–115. https://doi.org/10.1038/nature12496
Kaetzel CS (2014) Cooperativity among secretory IgA, the polymeric immunoglobulin receptor, and the gut microbiota promotes host–microbial mutualism. Immunol Lett 162(2):10–21. https://doi.org/10.1016/j.imlet.2014.05.008
Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K (2010) Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328(5986):1705–1709. https://doi.org/10.1126/science.1188454
Macpherson AJ, Köller Y, McCoy KD (2015) The bilateral responsiveness between intestinal microbes and IgA. Trends Immunol 36(8):460–470. https://doi.org/10.1016/j.it.2015.06.006
Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B (2015) Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 43(3): 541–553. https://doi.org/10.1016/j.immuni.2015.08.007
Pabst O, Cerovic V, Hornef M (2016) Secretory IgA in the coordination of establishment and maintenance of the microbiota. Trends Immunol 37(5):287–296. https://doi.org/10.1016/j.it.2016.03.002
Palm NW, De Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W (2014) Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158(5):1000–1010. https://doi.org/10.1016/j.cell.2014.08.006
Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, Hooper LV, Yarovinsky F (2014) Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 5(1):28–39. https://doi.org/10.4161/gmic.26489
Funding
The present study supported by the National Natural Science Foundation of China (31970503), the Chengdu Giant Panda Breeding Research Foundation (CPF2014-15), and the China Scholarship Council (201906910016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Statement
The experiment approved and supervised under the Institutional Animal Care and Use Committee of Sichuan Agricultural University (No. SYXKchuan 2014-187).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yi Zhoua, Xueqin Nia, and Ling Duan are Joint first authors
Supplementary information
ESM 1
(DOCX 411 kb)
Rights and permissions
About this article
Cite this article
Zhou, Y., Ni, X., Duan, L. et al. Lactobacillus plantarum BSGP201683 Improves the Intestinal Barrier of Giant Panda Microbiota-Associated Mouse Infected by Enterotoxigenic Escherichia coli K88. Probiotics & Antimicro. Prot. 13, 664–676 (2021). https://doi.org/10.1007/s12602-020-09722-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-020-09722-y