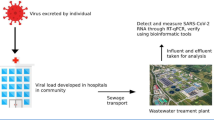
Abstract
The presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in wastewater has been reported as a result of fecal shedding of infected individuals. In this study, the occurrence of SARS-CoV-2 RNA was explored in primary-treated wastewater from two municipal wastewater treatment plants in Quintana Roo, Mexico, along with groundwater from sinkholes, a household well, and submarine groundwater discharges. Physicochemical variables were obtained in situ, and coliphage densities were determined. Three virus concentration methods based on adsorption-elution and sequential filtration were used followed by RNA isolation. Quantification of SARS-CoV-2 was done by RT-qPCR using the CDC 2020 assay, 2019-nCoV_N1 and 2019-nCoV_N2. The Pepper mild mottle virus, one of the most abundant RNA viruses in wastewater was quantified by RT-qPCR and compared to SARS-CoV-2 concentrations. The use of three combined virus concentration methods together with two qPCR assays allowed the detection of SARS-CoV-2 RNA in 58% of the wastewater samples analyzed, whereas none of the groundwater samples were positive for SARS-CoV-2 RNA. Concentrations of SARS-CoV-2 in wastewater were from 1.8 × 103 to 7.5 × 103 genome copies per liter (GC l−1), using the N1 RT-qPCR assay, and from 2.4 × 102 to 5.9 × 103 GC l−1 using the N2 RT-qPCR assay. Based on PMMoV prevalence detected in all wastewater and groundwater samples tested, the three viral concentration methods used could be successfully applied for SARS-CoV-2 RNA detection in further studies. This study represents the first detection of SARS-CoV-2 RNA in wastewater in southeast Mexico and provides a baseline for developing a wastewater-based epidemiology approach in the area.
Similar content being viewed by others
Introduction
The causal agent of the global pandemic of coronavirus disease (COVID-19) is the RNA enveloped severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that was first identified in Wuhan, China in 2019. This pandemic has caused over 120,383,919 positive cases and over 2,664,386 deaths around the world, Americas (North, Central, South) being the region with the highest number of confirmed positive cases of 53,160,109 (WHO, March 17, 2021). In Mexico the first case of COVID-19 was recorded on February 28 of 2020, and little more than one year later in March 16 of 2021, a total of 2,169,007 of accumulated confirmed positive cases have been reported, with 195,119 deaths (Secretaría de Salud, Gobierno de México (2021)).
The human-to-human transmission of SARS-CoV-2 occurs mainly through direct contact with an infected individual through droplets expelled by coughing, sneezing and respiratory droplets (Shereen et al., (Shereen et al. 2020)). In addition, a possible fecal–oral transmission route for SARS-CoV-2 has been proposed (Arslan et al., (Arslan et al. 2020)), due to the ability of the virus to replicate in enterocytes of the human intestine (Xiao et al., (Xiao et al. 2020)) resulting in high concentrations in feces of up to 108 genome copies per gram (Lescure et al., (Lescure et al. 2020); Pan et al., (Pan et al. 2020)), that can be released by fecal shedding of infected individuals (Chen et al., (Chen et al. 2020); Xing et al., (Xing et al. 2020); Lamers et al., (Lamers et al. 2020)). These high concentrations of SARS-CoV-2 can be found in stools of symptomatic and asymptomatic infected patients (Cheung et al., (Cheung et al. 2020)), resulting in the widespread presence of SARS-CoV-2 RNA in wastewater (Quilliam et al., (Quilliam et al. 2020)). To date, SARS-CoV-2 RNA has been reported in raw and treated wastewater in many countries around the world (Ahmed et al., (Ahmed et al. 2020); Fongaro et al., (Fongaro et al. 2020); Gerrity et al., (Gerrity et al. 2021); Gonzalez et al., (Gonzalez et al. 2020); Mlejnkova et al., (Mlejnkova et al. 2020)). Moreover, few studies have highlighted the risk of SARS-CoV-2 environmental contamination of water bodies that receive the discharge of treated or untreated wastewater (Lodder & de Roda Hudsman, (Lodder and Roda Husman 2020)).
Available data of the occurrence of SARS-CoV-2 RNA in wastewater can be used to implement a wastewater-based epidemiology (WBE) approach, which is a valuable tool to indirectly estimate viral prevalence in the population (Choi et al., (Choi et al. 2018); Rodriguez-Manzano et al., (Rodriguez-Manzano et al. 2010); Wu et al., (2020); Haramoto et al., (Haramoto et al. 2020)), and can also be used to identify new variants, if combined with sequence analysis. The advantages of WBE rely mainly on the possibility of early warning of disease outbreaks especially in certain areas where low prevalence or a decrease in prevalence occurs (Asghar et al., (Asghar et al. 2014); Hellmér et al., (Hellmér et al. 2014)). Under this scenario, the detection of SARS-CoV-2 in wastewater can be a feasible strategy to anticipate COVID-19 occurrence, because the viral concentration detected during different stages of wastewater treatment can be associated with the number of cases of COVID-19 reported in a particular area (Randazzo et al., (Randazzo et al. 2020)). As a result, the continuous surveillance of SARS-CoV-2 concentrations in wastewater can provide useful data for local authorities, which can lead to the implementation of rapid action that can prevent a negative impact on human and environmental health (McKinney et al., (McKinney et al. 2006); Casanova et al., (Casanova et al. 2009)). Therefore, WBE is a valuable strategy that could be implemented in countries such as Mexico, especially in areas with a high population increase rate or underprivileged societies with poor wastewater treatment and sanitation infrastructure.
To date, the presence of SARS-CoV-2 has not yet been reported in wastewater or in aquatic systems in Mexico, despite the growing concern of understanding the role and fate of this viral pathogen in the environment. In Mexico, Quintana Roo, located in the Yucatan Peninsula, has the highest population increase rate (3.5%) in the country (INEGI, (2020)), and is one of the most important touristic areas receiving, before the COVID-19 pandemic, more than 22 million visitors (Secretaría de Turismo, (2019)). Within the state, the municipality of Benito Juarez is where the highest total cumulative COVID-19 cases have been reported reaching 9,968, as of March 16, 2021. Since the first step to implement a successful WBE approach is to implement the methods for detection and quantification of SARS-CoV-2, the objective of this study was to determine the presence and concentration of SARS-CoV-2 RNA in wastewater and in groundwater in Quintana Roo, Mexico using three combined viral concentration methods and two qPCR assays.
Materials and Methods
Water Collection
Water samples were collected from August 2020 to January 2021 as follows:
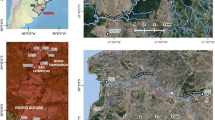
Wastewater. A total of 12 primary-treated wastewater samples (2 L) were collected from two wastewater treatment plants (WWTP), located in the city of Cancun, municipality of Benito Juarez (Fig. 1). Six samples (samples 24 to 29) were collected from the north WWTP (NT), which has an installed capacity of 350 L per second and treats 277 L per second using a dual treatment (biofilter and biological treatment). The NT WWTP serves 253,734 habitants, representing 28% of the total population of Cancun city of 888,797 (INEGI, (2020)), the major urban area in the Benito Juarez municipality. Also, six samples (samples 18 to 23) were collected from the northwestern WWTP (NP), which has an installed capacity of 200 L per second and treats 196 L per second using dual treatment. The NP WWTP serves 435,955 habitants, representing 34% of the population of Cancun city. Wastewater samples were collected by laboratory personnel and WWTP workers by using personal protective equipment (PPE) such as long pants, long sleeves, steel capped boots, protective vests, hard hats, safety glasses and face mask KN95. Samples were manually collected using a submersible in situ plastic sterile sampler by a grab sampling method (Haramoto et al., (Haramoto et al. 2020)).
Map showing the area of study, located in the state of Quintana Roo, Mexico. The colored dots correspond to the type of water collected as follows: blue = groundwater from sinkholes (cenotes), green = submarine groundwater discharges (SGD), red = groundwater from the household well, and orange = treated wastewater (before chlorination) from WWTP. The acronym of the site is shown per location (Color figure online)
Groundwater. All samples were collected using 10 L plastic sterile containers. Collection volumes were of 10–20 L (Table 1), depending on the physical characteristics of the collection site that allowed collecting sufficient water from the surface. A total of 10 samples were obtained from sinkholes located in the city of Cancun, Quintana Roo, and in the area locally known as Ruta de los Cenotes in Puerto Morelos, Quintana Roo (Fig. 1). Groundwater was also collected from one household well in the vicinity of Cancun city, which was sampled on two different dates. Also, five submarine groundwater discharges (SGDs) were collected parallel to the coast, within 50 km of the shoreline (Fig. 1), in the small fishermen town of Puerto Morelos, Quintana Roo. Certified divers using 10 L plastic containers and a submergible grab sampling strategy manually collected SGD samples.
Global Positioning System (GPS) coordinates were obtained from each field collection site (Table 1). Physicochemical variables of temperature (℃), conductivity (μS cm−1), total dissolved solids (mg l−1), dissolved oxygen (mg l−1) and pH were obtained in situ using a Hach HQd probe (Loveland, CO, USA). In SGD locations, temperature and conductivity were measured using the SBE19plus, CTD Sea Bird. All water samples were transported to the laboratory at room temperature for same day processing.
Virus Concentration Methods
A combination of previously published methods was used to concentrate viruses from groundwater and wastewater as follows:
Method A. Adsorption-elution followed by concentration using a centrifugal device (Centriprep® YM-50 filter). All water samples (1 to 29) were concentrated using this method. Adsorption-elution was conducted as reported previously by Katayama et al., ( (Katayama et al. 2002)). Briefly, a final concentration of 25 mM of MgCl2 was added to each water sample, and then the total volume was filtered through a cellulose-ester membrane (0.45 μm), using a vacuum pump until the membrane clogged. Cations were removed from the filter using 200 ml of H2SO4 0.5 M (pH 3.0), and the virus was eluted with 10 ml of NaOH 1 mM pH 10 in a centrifuge tube containing a neutralizing solution (50 μL of H2SO4 a 100 Mm and 100 μL of TE 1X). A second concentration step was done by using a Centriprep® YM-50 filter, according to Kitajima et al., (2012). Final volumes of 0.9–5.4 ml were obtained and stored at −20 °C.
Method B. Sequential filtration followed by polyethylene glycol (PEG) precipitation. A total of 16 samples were concentrated by this method, all 12 wastewater samples (18 to 29) and 4 groundwater samples (9 to 12). Briefly, water samples were filtered using a vacuum pump, and cellulose-ester membrane with different pore size, 0.8, 0.65, 0.45 and 0.22 µm. Water samples were sequentially filtered through each pore size. In each step, different volumes were filtered that ranged between 0.2 to 1.0 L. After sequential filtration a second concentration step was conducted, as previously reported by Hernández-Morga et al., ( (Hernández-Morga et al. 2009)) where polyethylene glycol (PEG) 8000 at 12%, and NaCl 0.8 mol l−1 were added to water concentrates followed by incubation at 4 ℃ for 12 h. After incubation, samples were centrifuged at 7000 rpm for 30 min at 4 ℃ twice. The supernatant was discarded, and the pellet was suspended in PBS buffer 1X. An equal volume of butanol–chloroform (1:1) was added to each sample and mixed manually, and centrifuged at 7000 rpm for 15 min (Hernández-Morga et al., 2009). The aqueous phase was recovered. The concentrated samples, at volumes of approximately 2.6–3.9 ml were stored at −20°C.
Method C. Sequential filtration followed using a centrifugal device (Centricon® plus 70 filter). A total of 8 samples were concentrated by this method, six wastewater samples (21, 22, 23, 27, 28 and 29) and two groundwater samples (9 and 11). Samples were concentrated following the sequential filtration as described in method B with different pore size cellulose-ester membrane. The second concentration step was carried out using Centricon® plus 70 filter, following manufacturer´s instructions. Samples were stored at -20º C. Final volumes of 0.6–1.3 ml were obtained and stored at −20 °C.
Somatic and Male F + Specific Coliphage Determination
Coliphage densities were determined from 10 ml of undiluted groundwater samples following the protocol described in Rosiles-González et al., ( (Rosiles-González et al. 2019)). Wastewater samples were serially diluted using a tenfold serial dilution of 100 to 10–4 in tryptone broth. Somatic and male F + specific coliphages were enumerated by a double layer plaque assay (U.S. Environmental Protection Agency method 1601) (U.S. EPA, (2015)) using ATCC strains 15,597 C-300 and 700,609 CN13 of Escherichia coli as hosts, respectively.
Nucleic Acid Isolation
Total RNA was isolated from 53 water concentrates obtained by the three viral concentration methods used in this study. Aliquots of 280 μl were used for RNA isolation, using QIAmp viral RNA mini kit (QIAGEN, Gilden, Germany), following manufacturer´s instructions. All samples were eluted in 50 μl AVE buffer and stored at −20℃.
Quantification of SARS-CoV-2 and PMMoV by RT-qPCR
Absolute quantification of SARS-CoV-2 and PMMoV was carried out by quantitative reverse transcription polymerase chain reaction (RT-qPCR) using purified total RNA in a 96 well-plate CFX96 (BioRad, Carlsbad, CA, USA) thermocycler. Total RNA was used as template for amplifications using the SuperScript III Platinum One-Step qRT-PCR System (Thermo Fischer Scientific, Waltham, MA, USA), following manufacturer`s instructions. The final volume of each reaction was of 25 μl containing: 5 µl of RNA, 12.5 µl of buffer 2x, 0.4 µl of MgSO4 (50 mM), 1.25 µl of the mix primers/TaqMan probe (5.0 y 2.5 nmol, respectively) and 0.5 µl SuperScript® III/Platinum® Taq. Negative controls were prepared using 51 of PCR grade water as template. A standard curve was determined for each RT-qPCR assay by using a tenfold serial dilution of 100 to 105 DNA genome copies DNA control for SARS-CoV-2 (Integrated DNA Technologies, Coralville, IA, USA) and a plasmid DNA control constructed at the laboratory for PMMoV (Rosiles-González et al., (Rosiles-González et al. 2017)). Each standard curve was established based on the linear relationship between the log initial concentration of plasmid DNA and the threshold cycle (Ct) value. Viral quantification was determined at genome copies per liter (GC l−1), and the equivalent volume of water tested ranged from 1.8 to 121 ml (method A), 1.4 to 9.3 ml (method B), and from 5.7 to 15.4 ml (method C). Samples were considered positive only when 10 or more copies of the viral genome were registered. Primers and probes used for SARS-CoV-2 detection were obtained from Integrated DNA Technologies (IDT, Coralville, IA, USA), using the 2019-nCoV CDC EUA Kit and the 2019-nCoV_N Positive. The thermal cycling conditions of the qPCR assays were as follows: initial incubation at 50 °C for 15 min and initial denaturation at 95 °C for 2 min, 45 cycles at 95 °C for 15 s and 55 °C for 30 s, for N1 and N2 genes. For PMMoV thermal cycling conditions were the same except for alignment temperature which was of 60 °C.
Results
Physicochemical Variables
The average minimum temperature of 26.9 ± 1.6 °C was recorded in the household well, whereas the average maximum temperature of 29.3 ± 1.1 °C was recorded in primary-treated wastewater (Table 2). The average minimum conductivity of 953.5 ± 241 μS cm−1 was recorded in groundwater from sinkholes, whereas the average maximum of 43,800 ± 6.7 μS cm−1 was registered in SGDs. The average minimum of total dissolved solids of 444 ± 125 mg l−1 was registered in groundwater from sinkholes, and the maximum value of 21,500 mg l−1 was recorded in SGD (Table 2). Dissolved oxygen average minimum concentrations of 1.3 ± 1.7 mg l−1 were registered in primary-treated wastewater, and the maximum value of 4.7 mg l−1 was recorded from SGDs (Table 2). All samples showed pH values higher than 6.9 (alkaline sites); however, the lowest pH average value of 6.9 ± 0.05 was registered from the household well, and the highest average values of 7.7 ± 0.4 were detected in primary-treated wastewater (Table 2).
Densities of Male F + Specific and Somatic Coliphages
Male F + specific coliphages were detected in all wastewater samples, and in 17% of groundwater samples from sites 5, and 8, that corresponded to groundwater from sinkholes, and site 13 that corresponded to a SGD. The concentration male F + specific coliphages ranged from 5.0 × 103 to 1.1 × 107 plaque forming units per 100 ml (PFU 100 ml−1) in primary-treated wastewater, and from 1.0 × 101 to 8.0 × 101 PFU 100 ml−1 in groundwater (Table 3). Somatic coliphages were detected in all primary-treated wastewater samples, and in 23% of groundwater samples in sites 2, 4 and 8, which corresponded to groundwater from sinkholes, and site 13 corresponding to a SGD. The concentration of somatic coliphages ranged from 6.0 × 103 to 1.1 × 107 PFU 100 ml−1 in treated wastewater, and from 2.0 × 101 to 8.0 × 101 in groundwater (Table 3). From all samples, the highest densities of male F + specific coliphages and somatic coliphages were recorded in sample 23 that corresponded to primary-treated wastewater collected from the northwestern (NP) WWTP on November 24, 2020 (Table 3).
Occurrence of SARS-CoV-2
Primary-treated wastewater samples were positive for the detection of SARS-CoV-2 by using both RT-qPCR N1 and N2 assays, whereas viral RNA was not detected in any of the groundwater samples (Table 4). The RT-qPCR amplification of the N1 gene fragment allowed the detection of SARS-CoV-2 in 25% (3/12) of the primary-treated wastewater samples that were processed by the viral concentration method A, and in 16% (2/12) of the primary-treated wastewater samples processed using the viral concentration method B (Table 4). Also, RT-qPCR amplification of the N2 gene fragment allowed the detection of SARS-CoV-2 in 33% (4/12) of the primary-treated wastewater samples that were processed by the viral concentration method A, and in 25% (3/12) of the samples processed using viral concentration method B (Table 4).
Even though only a subset of primary-treated wastewater samples was processed by the viral concentration method C, results showed that at least 33% (2/6) of the samples tested were positive for SARS-CoV-2 using this method together with the RT-qPCR N1 assay. Moreover, the RT-qPCR N2 assay in combination with the viral concentration method C allowed the detection of SARS-CoV-2 in 50% (3/6) of the primary-treated wastewater samples tested (Table 4). Overall, results showed that the RT-qPCR assays allowed the detection of the N1 and N2 fragment simultaneously in samples 19 and 23 (method A), sample 20 (method B) and samples 21 and 23 (methods A and C) (Table 4). The use of the three combined viral concentration methods A, B and C, together with two qPCR assays, allowed the detection of SARS-CoV-2 RNA in 58% (7/12) of the primary-treated wastewater samples tested.
Wastewater samples tested in this study were all positive for the amplification of PMMoV by RT-qPCR (Table 4). These results showed that inhibition of the RT-qPCR reaction was absent. However, not all of the wastewater samples were positive for SARS-CoV-2, the prevalence of positive SARS-CoV-2 samples that tested positive for at least one SARS-CoV-2 RT-qPCR target was different depending on the WWTP from where the sample was collected. All of the samples collected at the WWTP NP (samples 18 to 23) were positive to at least one SARS-CoV-2 RT-qPCR target (Table 4). Whereas wastewater samples collected from the WWTP NT were all negative with the only exception of sample 25 that was positive to at least one SARS-CoV-2 RT-qPCR target (Table 4).
The concentrations of SARS-CoV-2 obtained by the RT-qPCR N1 assay ranged from 1.8 × 103 GC l−1 in sample 21 (method C) to 7.5 × 103 GC l−1 in sample 18 (method A); whereas the concentrations of SARS-CoV-2 obtained by the RT-qPCR N2 assay ranged from 2.4 × 102 GC l−1 in sample 22 (method C) to 5.9 × 103 GC l−1 in sample 23 (method A) (Table 4). The highest concentration of SARS-CoV-2 was recorded in sample 18 (Table 4), which was collected from the northwestern (NP) WWTP on September 29 of 2020 (Table 1).
When SARS-CoV-2 RNA was detected, the cumulative COVID-19 cases reported in the Benito Juarez municipality, were within the range of 5,640 to 6,668 (September 29 to November 24, 2020). Results showed that SARS-CoV-2 qPCR signals were detected in a total of five samples by the RT-qPCR N1 assay, and in six samples with the RT-qPCR N2 assay, in dates where a range of COVID-19 cases per day were reported to be from 10 to 34 (Fig. 2).
Occurrence of PMMoV
All the groundwater (17/17) and wastewater (12/12) samples that were processed by the method A tested positive for the detection of PMMoV by RT-qPCR (Table 4). In addition, PMMoV was detected in all samples tested from the subset of groundwater and wastewater samples that were processed by the viral concentration methods B and C (Table 4). The concentrations of PMMoV in groundwater were from 3.2 × 102 GC l−1 in sample 14 (method A), to 2.0 × 105 GC l−1 in sample 11 (method C); whereas in groundwater PMMoV concentrations were higher from 1.2 × 107 GC l−1 in sample 29 (method A), to 2.4 × 109 in sample 23 (method B).
Discussion
This study constitutes the first surveillance of SARS-CoV-2 RNA in primary-treated wastewater and groundwater in Mexico by using three viral concentration methods (based on adsorption-elution and sequential filtration) and two RT-qPCR assays. The occurrence of SARS-CoV-2 RNA in raw and treated wastewater has been reported in different countries around the world (Ahmed et al., (Ahmed et al. 2020); Haramoto et al., (Haramoto et al. 2020); La Rosa et al., (2020); Randazzo et al., (Randazzo et al. 2020)). Even though, one standardized method that can be applied elsewhere might not be feasible, the concentration and recovery of SARS-CoV-2 has been successfully achieved by using common methods applied in the study of viruses in water such as adsorption-elution (Haramoto et al., (Haramoto et al. 2020)), ultrafiltration (Philo et al., (Philo et al. 2021); Sherchan et al., (Sherchan et al. 2020)), and polyethylene glycol (PEG) precipitation (Philo et al., 2020). In addition, there are different RT-qPCR assays available for the successful detection of SARS-CoV-2 RNA in wastewater (Ahmed et al., (Ahmed et al. 2020); Corman et al., (Corman et al. 2020); CDC, (2020)). However, in Mexico the presence of SARS-CoV-2 in wastewater and its fate in the environment is still unknown. Thus, results from this study provide a baseline for the use of at least three viral concentration methods that could be applied for the detection of SARS-CoV-2 RNA by two RT-qPCR assays in wastewater in Mexico. Moreover, the use in this study of PMMoV, a viral fecal indicator (Kitajima et al., (Kitajima et al. 2018)), demonstrated a high performance of methods A, B and C, supporting their applicability for SARS-CoV-2 RNA detection in further studies.
The fate of SARS-CoV-2 in the environment is not yet well understood, in fact, there are very few studies where the presence of SARS-CoV-2 has been investigated in superficial water bodies that receive raw or treated wastewater and combined sewage overflows (Rimoldi et al., (2020)). To date there is an urgent need for further assessment of the fate of SARS-CoV-2 in the environment and the potential risk for public and environmental health, especially in low sanitation areas (Guerrero-Latorre et al., (Guerrero-Latorre et al. 2020)). In this study, groundwater was collected from sinkholes or cenotes in the northeast karst aquifer of the Yucatan peninsula. Sinkholes are filled with water from the subterranean karst aquifer that is vulnerable to fecal contamination, as a result of the discharge of treated and non-treated wastewater, failing or inadequate sewage systems, septic tanks that are not well kept and sewage overflow (Metcalfe et al., (Metcalfe et al. 2020); Marín et al., (Marín et al. 2000)). In addition, submarine groundwater discharges were collected in coastal areas since SGDs can be a source of fecal material (Kantú-Manzano et al., (Kantú-Manzano et al. 2018)). In this study, results showed that all groundwater samples were negative for the detection of SARS-CoV-2 RNA by RT-qPCR N1 and N2 assays, even though, the Pepper mild mottle virus, a viral indicator of fecal contamination was detected in all groundwater samples tested. Moreover, previous studies conducted in groundwater from sinhkoles in the area, have demonstrated the presence of enteric viruses such as human adenoviruses and noroviruses (Rosiles-González et al., (Rosiles-González et al. 2019)), indicating that viral fecal contamination is reaching the aquifer as a result of poor sanitation. Although the environmental transmission of SARS-CoV-2 has not been confirmed and the viral RNA was not present in groundwater, it is well known that the virus can persist in wastewater (Barcelo, (Barcelo 2020)), thus, the potential health risks derived from the direct discharge of non-treated or inadequately treated wastewater into groundwater needs to be considered in further research.
In this study, SARS-CoV-2 RNA was detected in 58% of the total primary-treated wastewater samples (7/12), in concentrations of 102 to 103 GC l−1. To date, different data is available as a result of the continuous monitoring of SARS-CoV-2 in wastewater (Ahmed et al., (Ahmed et al. 2020); Haramoto et al., (Haramoto et al. 2020); Lodder & de Roda Husman, (Lodder and Roda Husman 2020); Wu et al., (2020); Wurtzer et al., (2020)), however, continuous surveillance in different geographical regions is recommended (Kitajima et al., (Kitajima et al. 2020)), since the prevalence and concentration of SARS-CoV-2 RNA in wastewater can be very variable. For example, in treated wastewater the prevalence of SARS-CoV-2 RNA can be as high as 75% (Wurtzer et al., (2020)) or as low as 20% in secondary treated wastewater (Haramoto et al., (Haramoto et al. 2020)). In addition, SARS-CoV-2 RNA copies per liter can span a wide range as shown in non-treated wastewater where concentrations can be within a range of 102 to 106 (Ahmed et al., (Ahmed et al. 2020); Corpuz et al., (Corpuz et al. 2020); Wurtzer et al., (2020)), whereas in treated wastewater concentrations can be up to 105 GC l−1 (Haramoto et al., (Haramoto et al. 2020); Randazzo et al., (Randazzo et al. 2020); Wurtzer et al., (2020)). Even though a wide range of SARS-CoV-2 concentrations has been reported to occur in wastewater, it is important to consider that in this study plasmids were used as calibration standards, therefore, the efficiency of the reverse transcription was not considered, which could lead to an underestimation of the real SARS-CoV-2 concentrations in the original wastewater samples. In addition, viral concentrations were calculated in the original wastewater sample assuming no loss of virus during the steps of the detection procedure, which can also result in underestimation of the real viral concentrations (Haramoto et al., (Haramoto et al. 2020)). In tropical areas, such as the southeast of Mexico, information regarding the different methods for SARS-CoV-2 RNA detection in wastewater and its implications are scarce. Therefore, it is important to apply molecular epidemiology methodologies and to conduct the surveillance of SARS-CoV-2 in wastewater and in receiving environmental water.
The wastewater-based epidemiology (WBE) is a valuable approach that allows the monitoring of COVID-19 incidence in communities and provides useful information to local authorities for the implementation of public health policies (Hart & Halden, (Hart and Halden 2020)). The correspondence of SARS-CoV-2 concentrations in wastewater with the peaks of new cases of COVID-19 infection have been observed previously, indicating that this approach can be successfully used for COVID-19 monitoring (Gonzalez et al., (Gonzalez et al. 2020); Haramoto et al., (Haramoto et al. 2020)). This study was conducted in Quintana Roo, a state located in the Yucatan peninsula of Mexico, which is characterized by high temperatures, low annual rainfall, and the presence of extreme climatological events such as tropical storms and hurricanes (Metcalfe et al., (Metcalfe et al. 2020)). Wastewater sampling was conducted in WWTPs from the municipality of Benito Juarez within a short three-month span, from September to November 2020, when two hurricanes impacted the area (Hurricanes Delta and Zeta), with the consequent sewage overflow and extreme flooding events. Therefore, the highest concentrations of SARS-CoV-2 RNA detected did not corresponded exactly with the highest number of 34 COVID-19 new cases per day that were reported for the Municipality of Benito Juarez within the three-month span of this study. Nonetheless, without extreme climatic events, the methodologies implemented in this study allowed the detection of SARS-CoV-2 RNA with as low as 10 new COVID-19 cases per day. WBE approach could be applied in Mexico and in other tropical areas through the development of methodologies to correlate the viral concentrations and the incidence of the disease, especially in low sanitation areas, or areas where extensive testing of symptomatic individuals is not possible. Furthermore, continuous monitoring of the environmental fate of SARS-CoV-2 should be conducted in this, and in other karst aquifers that are vulnerable to viral contamination.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O’Brien, J. W., Choi, P. M., Kitajima, M., Simpson, S. L., Li, J., Tscharke, B., Verhagen, R., Smith, W. J. M., Zaugg, J., Dierens, L., Hugenholtz, P., Thomas, K. V., & Mueller, J. F. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Science of the Total Environment. https://doi.org/10.1016/j.scitotenv.2020.138764
Arslan, M., Xu, B., & Gamal, & El-Din, M. (2020). Transmission of SARS-CoV-2 via fecal-oral and aerosols–borne routes: Environmental dynamics and implications for wastewater management in underprivileged societies. Science of the Total Environment, 743, 140709. https://doi.org/10.1016/j.scitotenv.2020.140709
Asghar, H., Diop, O. M., Weldegebriel, G., Malik, F., Shetty, S., El Bassioni, L., Akande, A. O., Al Maamoun, E., Zaidi, S., Adeniji, A. J., Burns, C. C., Deshpande, J., Oberste, S. M., & Lowther, S. A. (2014). Environmental surveillance for polioviruses in the global polio eradication initiative. The Journal of Infectious Diseases, 210(Suppl 1), S294–S303. https://doi.org/10.1093/infdis/jiu384
Barcelo, D. (2020). An environmental and health perspective for COVID-19 outbreak: meteorology and air quality influence, sewage epidemiology indicator, hospitals disinfection, drug therapies and recommendations. Journal of Environmental Chemical Engineering, 8, 104006. https://doi.org/10.1016/j.jece.2020.104006
Casanova, L., Rutala, W. A., Weber, D. J., & Sobsey, M. D. (2009). Survival of surrogate coronaviruses in water. Water Research, 43, 1893–1898. https://doi.org/10.1016/j.watres.2009.02.002
CDC (2020, January 01). CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. Retrieved June 3, 2020, from https://www.fda.gov/media/134922/download
Chen, Y., Chen, L., Deng, Q., Zhang, G., Wu, K., Ni, L., Yang, Y., Liu, B., Wang, W., Wei, C., Yang, J., Ye, G., & Cheng, Z. (2020). The presence of SARS-CoV-2 RNA in feces of COVID-19 patients. Journal of Medical Virology. https://doi.org/10.1002/jmv.25825
Cheung, K. S., Hung, I. F., Chan, P. P., Lung, K., Tso, E., Liu, R., Ng, Y., Chu, M. Y., Chung, T. W., Tam, A. R., Yip, C. C., Leung, K.-H., Yim-Fong Fung, A., Zhang, R. R., Lin, Y., Cheng, H. M., Zhang, A. J., To, K. K., Chan, K.-H., … Leung, W. K. (2020). Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology, 159, 81–95. https://doi.org/10.1053/j.gastro.2020.03.065
Choi, P. M., O’Brien, J. W., Li, J., Jiang, G., Thomas, K. V., & Mueller, J. F. (2018). Population histamine burden assessed using wastewater-based epidemiology: the association of 1,4-methylimidazole acetic acid and fexofenadine. Environment International, 120, 172–180. https://doi.org/10.1016/j.envint.2018.08.009
Corman, V. M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D. K., Bleicker, T., Brünink, S., Schneider, J., Schmidt, M. L., Mulders, D. G., Haagmans, B. L., & van DV., van den Brink S., Wijsman, L., Goderski, G., Romette, JL., Ellis, J., Zambon, M., Peiris, M., Goossens, H., Reusken, C., Koopmans, MP., & Drosten, C. . (2020). Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance, 25(3), 2000045. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
Corpuz, M. V. A., Buonerba, A., Vigliotta, G., Zarra, T., Ballesteros, F., Campiglia, P., Belgiorno, V., Korshin, G., & Naddeo, V. (2020). Viruses in wastewater: occurrence, abundance and detection methods. Science of the Total Environment, 745, 140910. https://doi.org/10.1016/j.scitotenv.2020.140910
Fongaro, G., Stoco, P. H., Sobral, Marques Souza, & D., Grisars, E. C., Magri, M. E., Rogovski, P., Schörner, A. M., Barazzetti, F. H., Christoff, A. P., de Oliveira, L. F. V., Bazzo, M. L., Wagner, G., Hernández, M., & Rodriguez-Lázaro, D. (2020). SARS-CoV-2 in human sewage in Santa Catalina Brazil. medRxiv. https://doi.org/10.1101/2020.06.26.20140731
Gerrity, D., Papp, K., Stoker, M., Sims, A., & Frehner, W. (2021). Early-pandemic wastewater surveillance of SARS-CoV-2 in southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Research, 10, 100086. https://doi.org/10.1016/j.wroa.2020.100086
Gonzalez, R., Curtis, K., Bivins, A., Bibby, K., Weir, M., Yetka, K., Thompson, H., Keeling, D., Mitchell, J., & Gonzalez, D. (2020). COVID-19 surveillance in southeastern virginia using wastewater -based epidemiology. Water Research, 186, 116296. https://doi.org/10.1016/j.watres.2020.116296
Guerrero-Latorre, L., Ballesteros, I., Villacrés-Granda, I., Granda, M. G., Freire-Paspuel, B., & Ríos-Touma, B. (2020). SARS-CoV-2 in river water: implications in low sanitation countries. Science of the Total Environment, 743, 140832. https://doi.org/10.1016/j.scitotenv.2020.140832
Haramoto, E., Malla, B., Thakali, O., & Kitajima, M. (2020). First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Science of the Total Environment. https://doi.org/10.1016/j.scitotenv.2020.140405
Hart, O. E., & Halden, R. U. (2020). Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Science of the Total Environment, 730, 138875. https://doi.org/10.1016/j.scitotenv.2020.138875
Hellmér, M., Paxéus, N., Magnius, L., Enache, L., Arnholm, B., Johansson, A., Bergström, T., & Norder, H. (2014). Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Applied and Environmental Microbiology, 80, 6771–6781. https://doi.org/10.1128/AEM.01981-14
Hernández-Morga, J., Leon, F. J., Peraza, G. F., Gil, S. B. G., & Chaidez, C. (2009). Detection and characterization of Hepatitis A Virus and Norovirus present in estuarine water samples using ultrafiltration- RT-PCR integrated methods. Journal of Applied Microbiology, 106(5), 1579–1590. https://doi.org/10.1111/j.1365-2672.2008.04125.x
INEGI. (2020). Inicio. México en cifras. Quintana Roo. Instituto Nacional de Estadística y Geografía. México. Retrieved January, 2021, from https://www.inegi.org.mx/app/areasgeograficas/
Kantú-Manzano, C., Herrera-Silveira, J., & Arcega-Cabrera, F. (2018). Influence of coastal submarine groundwater discharges on seagrass communities in a subtropical karstic environment. Bulletin of Environmental Contamination and Toxicology, 100, 176–183. https://doi.org/10.1007/s00128-017-2259-3
Katayama, H., Shimasaki, A., & Ohgaki, S. (2002). Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Applied and Environmental Microbiology, 68(3), 1033–1039. https://doi.org/10.1128/aem.68.3.1033-1039.2002
Kitajima, M., Haramoto, E., Phanuwan, C., Katayama, H., & Furumai, H. (2012). Molecular detection and genotyping of human noroviruses in influent and effluent water at a wastewater treatment plant in Japan. Journal of Applied Microbiology, 112(3), 605–613. https://doi.org/10.1111/j.1365-2672.2012.05231.x
Kitajima, M., Sassi, H. P., & Torrey, J. R. (2018). Pepper mild mottle virus as a water quality indicator. Npj Clean Water, 1, 19. https://doi.org/10.1038/s41545-018-0019-5
Kitajima, M., Ahmed, W., Bibby, K., Carducci, A., Gerba, C. P., & Hamilton, K. A. (2020). SARS-CoV-2 in wastewater: state of the knowledge and research needs. Science of the Total Environment. https://doi.org/10.1016/j.scitotenv.2020.139076
La Rosa, G., Iaconelli, M., Mancini, P., Bonanno, F. G., Veneri, C., Bonadonna, L., Lucentini, L., Suffredini, E. (2020). First detection of SARS-CoV-2 in untreated wastewaters in Italy. medRxiv. https://doi.org/10.1101/2020.04.25.20079830
Lamers, M. M., Beumer, J., van der Vaart, J., Knoops, K., Puschhof, J., Breugem, T. I., Ravelli, R. B. G., Paul van Schayck, J., Mykytyn, A. Z., Duimel, H. Q., van Donselaar, E., Riesebosch, S., Kuijpers, H. J. H., Schipper, D., van de Wetering, W., de Graaf, M., Koopmans, M., Cuppen, E., Peters, P. J., … Clevers, H. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science. https://doi.org/10.1126/science.abc1669
Lescure, F.-X., Bouadma, L., Nguyen, D., Parisey, M., Wicky, P.-H., Behillil, S., Gaymard, A., Bouscambert-Duchamp, M., Donati, F., Le Hingrat, Q., Enouf, V., Houhou-Fidouh, N., Valette, M., Mailles, A., Lucet, J.-C., Mentre, F., Duval, X., Descamps, D., Malvy, D., … van-der-Werf, S., & Yazdanpanah, Y. . (2020). Clinical and virological data of the first cases of COVID-19 in Europe: a case series. The LAncet Infectious Diseases. https://doi.org/10.1016/S1473-3099(20)30200-0
Lodder, W., & de Roda Husman, A. M. (2020). SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterology and Hepatology, 5(6), 533–534. https://doi.org/10.1016/S2468-1253(20)30087-X
Marín, L. E., Steinich, B., Pacheco, J., & Escolero, O. A. (2000). Hydrogeology of a contaminated sole-source karst aquifer, Mérida, Yucatán. Mexico. Geofísica Internacional, 49(4), 359–365.
McKinney, K. R., Gong, Y. Y., & Lewis, T. G. (2006). Environmental transmission of SARS at Amoy Gardens. Journal of Environmental Health, 68, 26–30.
Metcalfe, S. E., Schmook, B., Boyd, D. S., de la Barreda-Bautista, B., Endfield, G. E., Mardero, S., Manzón, M. C., Medina, R. G., Munguia, G. M. T., & Olmedo, S. N. (2020). Community perception, adaptation and resilience to extreme weather in the Yucatan Peninsula. Mexico. Regional Environmental Change, 20(1), 1–15. https://doi.org/10.1007/s10113-020-01586-w
Mlejnkova, H., Sovova, K., Vasickova, P., Ocenaskova, V., Jasikova, L., & Juranova, E. (2020). Preliminary study of Sars-Cov-2 occurrence in wastewater in the Czech republic. International Journal of Environmenatl Research and Public Health, 17(15), 5508. https://doi.org/10.3390/ijerph17155508
Pan, Y., Zhang, D., Yang, P., Poon, L. L. M., & Wang, Q. (2020). Viral load of SARS-CoV-2 in clinical samples. The Lancet Infectious Diseases, 20, 411–412. https://doi.org/10.1016/S1473-3099(20)30113-4
Philo, S. E., Keim, E. K., Swanstrom, R., Ong, A. Q. W., Burnor, E. A., Kossik, A. L., Harrison, J. C., Demeke, B. A., Zhou, N. A., Beck, N. K., Shirai, J. H., & Meschke, J. S. (2021). A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Science of the Total Environment, 760, 144215. https://doi.org/10.1016/j.scitotenv.2020.144215
Quilliam, R. S., Weidmann, M., Moresco, V., Purshouse, H., O’Hara, Z., & Oliver, D. M. (2020). COVID-19: the environmental implications of shedding SARS-CoV-2 in human faeces. Environment International, 140, 105790. https://doi.org/10.1016/j.envint.2020.105790
Randazzo, W., Truchado, P., Cuevas-Ferrando, E., Simón, P., Allende, A., & Sánchez, G. (2020). SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Research. https://doi.org/10.1016/j.watres.2020.115942
Rimoldi, S. G., Stefani, F., Gigantiello, A., Polesello, S., Comandatore, F., Mileto, D., Maresca, M., Longobardi, C., Mancon, A., Romeri, F., Pagani, C., Moja, L., Gismondo, M. R., & Salerno, F. (2020). Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. medRxiv. https://doi.org/10.1101/2020.05.01.20086009
Rodriguez-Manzano, J., Miagostovich, M., Hundesa, A., Clemente-Casares, P., Carratala, A., Buti, A., Jardí, R., & Girones, R. (2010). Analysis of the evolution in the circulation of HAV and HEV in Eastern Spain by testing urban sewage samples. Journal of Water and Health, 8(2), 346–354. https://doi.org/10.2166/wh.2009.042
Rosiles-González, G., Ávila-Torres, G., Moreno-Valenzuela, O. A., Acosta-González, G., Leal-Bautista, R. M., Grimaldo-Hernández, C. D., Brown, J. K., Chaidez-Quiroz, C., Betancourt, W. Q., Gerba, C. P., & Hernández-Zepeda, C. (2017). Occurrence of Pepper mild mottle virus (PMMoV) in groundwater from a karst aquifer system in the Yucatan Peninsula. Mexico. Food and Environmental Virology, 9(4), 487–497. https://doi.org/10.1007/s12560-017-9309-1
Rosiles-González, G., Ávila-Torres, G., Moreno-Valenzuela, O. A., Chaidez-Quiroz, C., Hernández-Flores, C. I., Acosta-González, G., Brown, J. K., Betancourt, W. Q., Gerba, C. P., & Hernández-Zepeda, C. (2019). Norovirus and human adenovirus occurrence and diversity in recreational water in a karst aquifer in the Yucatan Peninsula. Mexico. Journal of Applied Microbiology, 127(4), 1255–1269. https://doi.org/10.1111/jam.14385
Sherchan, S. P., Shahin, S., Ward, L. M., Tandukar, S., Aw, T. G., Schmitz, B., Ahmed, W., & Kitajima, M. (2020). First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Science of the Total Environment. https://doi.org/10.1016/j.scitotenv.2020.140621
Shereen, M. A., Khan, S., Kazmi, A., Bashir, N., & Siddique, R. (2020). COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research, 24, 91–98. https://doi.org/10.1016/j.jare.2020.03.005
Secretaría de Turismo. (2019, December). Quintana Roo ¿Cómo vamos en turismo de Quintana Roo? Retrieved February 10, 2021, from http://sedeturqroo.gob.mx/ARCHIVOS/como_vamos_enero_octubre_2018.pdf.
Secretaría de Salud, Gobierno de México. (2021, February). Informe Técnico Diario COVID-19 México. Retrieved February 10, 2021, from https://www.gob.mx/salud/documentos/coronavirus-covid-19-comunicado-tecnico-diario-238449
U.S. EPA. (2015, April 17). Review of coliphages as possible indicators of fecal contamination for ambient water quality. EPA 820-R15–098. Washington, DC: Office of Water, Science and Technology Health and Ecological Criteria. Division. Retrieved March 30, 2017, from https://www.epa.gov/sites/production/files/2016-07/documents/review_of_coliphages_as_possible_indicators_of_fecal_contamination_for_ambient_water_quality.pdf.
WHO. (2020, February 18). Coronavirus Disease (COVID-19) Press Conference 18 February 2020. Retrieved February 18, 2020, from https://www.who.int/docs/default-source/coronaviruse/transcripts/who-audio-emergencies-coronavirus-full-press-conference-18feb2020-final.pdf?sfvrsn=5209d6c3_2.
Wu, F., Xiao, A., Zhang, J., Gu, X., Lee, W., Kauffman, K., Hanage, W., Matus, M., Ghaeli, N., Endo, N., Duvallet, C., Moniz, K., Erickson, T., Chai, P., Thompson, J., & Alm, E. (2020). SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. https://doi.org/10.1101/2020.04.05.20051540
Wurtzer, S., Marechal, V., Mouchel, J. M., Maday, Y., Teyssou, R., Richard, E., Almayrac, J. L., & Moulin, L. (2020). Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral 621 genome quantification in Paris wastewaters. medRxiv. https://doi.org/10.1101/2020.04.12.20062679
Xiao, F., Tang, M., Zheng, X., Liu, Y., Li, X., & Shan, H. (2020). Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology, 158, 1831–1833. https://doi.org/10.1053/j.gastro.2020.02.055
Xing, Y., Ni, W., Wu, Q., Li, W., Li, G., Tong, J., Song, X.-F., Wing-Kin Wong, G., & Xing, Q.-S. (2020). Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. Journal of Microbiology, Immunology and Infection, 53(3), 473–480. https://doi.org/10.1016/j.jmii.2020.03.021
Acknowledgements
The authors want to give special thanks to Dr. Antonio Almazán Becerril, M.C. Benjamín Delgado Pech and M.C. Jorge Carlos Peniche Pérez for their support in field collection of water form submarine groundwater discharges. The authors also want to give special thanks to AGUAKAN S.A. de C.V. for their collaboration during wastewater collection and for providing important data of the wastewater treatment plants. This project (Grant number 313216) was funded by Consejo Nacional de Ciencia y Tecnología (Conacyt)/“Apoyo para Proyectos de Investigación Científica, Desarrollo Tecnológico e Innovación en Salud ante la Contingencia por COVID-19.”
Funding
This project (Grant number 313216) was funded by Consejo Nacional de Ciencia y Tecnología (Conacyt)/ “Apoyo para Proyectos de Investigación Científica, Desarrollo Tecnológico e Innovación en Salud ante la Contingencia por COVID-19”.
Author information
Authors and Affiliations
Contributions
GRG: Methodology, Validation, Investigation, Formal analysis. VHCJ: Methodology, Investigation, Formal analysis. OAV: Writing-review & editing. WQB: Methodology, Writing-review & editing. CPG: Methodology, Writing-review & editing. RTT: Resources, Writing-review & editing. LAG: Funding acquisition, Writing-review & editing. CHZ: Conceptualization, Funding acquisition, Project Administration, Methodology, Investigation, Resources, Supervision, Writing- original draft, Formal analysis.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rosiles-González, G., Carrillo-Jovel, V.H., Alzate-Gaviria, L. et al. Environmental Surveillance of SARS-CoV-2 RNA in Wastewater and Groundwater in Quintana Roo, Mexico. Food Environ Virol 13, 457–469 (2021). https://doi.org/10.1007/s12560-021-09492-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-021-09492-y