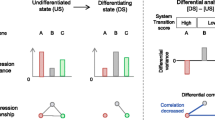
Abstract
Deciphering regulatory patterns of neural stem cell (NSC) differentiation with multiple stages is essential to understand NSC differentiation mechanisms. Recent single-cell transcriptome datasets became available at individual differentiation. However, a systematic and integrative analysis of multiple datasets at multiple temporal stages of NSC differentiation is lacking. In this study, we propose a new method integrating prior information to construct three gene regulatory networks at pair-wise stages of transcriptome and apply this method to investigate five NSC differentiation paths on four different single-cell transcriptome datasets. By constructing gene regulatory networks for each path, we delineate their regulatory patterns via differential topology and network diffusion analyses. We find 12 common differentially expressed genes among the five NSC differentiation paths, with one common regulatory pattern (Gsk3b_App_Cdk5) shared by all paths. The identified regulatory pattern, partly supported by previous experimental evidence, is essential to all differentiation paths, but it plays a different role in each path when regulating other genes. Together, our integrative analysis provides both common and specific regulatory mechanisms for each of the five NSC differentiation paths.
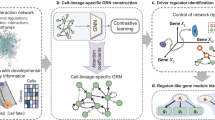
Graphical Abstract









Similar content being viewed by others
References
Schwartz PH, Brick DJ, Stover AE, Loring JF, Muller FJ (2008) Differentiation of neural lineage cells from human pluripotent stem cells. Methods 45(2):142–158. https://doi.org/10.1016/j.ymeth.2008.03.007
English D, Sanberg PR (2006) Neural specification of stem cell differentiation. Stem Cells Dev 15(2):139–140. https://doi.org/10.1089/scd.2006.15.139
Qiao SP, Liu Y, Han FT, Guo M, Hou XL, Ye KR, Deng S, Shen YJ, Zhao YF, Wei HY, Song B, Yao LF, Tian WM (2018) An intelligent neural stem cell delivery system for neurodegenerative diseases treatment. Adv Healthc Mater. https://doi.org/10.1002/adhm.201800080
Hashimshony T, Wagner F, Sher N, Yanai I (2012) CEL-Seq: single-cell RNA-seq by multiplexed linear amplification. Cell Reports 2(3):666–673. https://doi.org/10.1016/j.celrep.2012.08.003
Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R (2013) Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10(11):1096–1098. https://doi.org/10.1038/NMETH.2639
Pina C, Teles J, Fugazza C, May G, Wang D, Guo Y, Soneji S, Brown J, Eden P, Ohlsson M, Peterson C, Enver T (2015) Single-cell network analysis identifies DDIT3 as a nodal lineage regulator in hematopoiesis. Cell Rep 11(10):1503–1510. https://doi.org/10.1016/j.celrep.2015.05.016
Hu Y, Hase T, Li HP, Prabhakar S, Kitano H, Ng SK, Ghosh S, Wee LJ (2016) A machine learning approach for the identification of key markers involved in brain development from single-cell transcriptomic data. BMC Genomics 17(Suppl 13):1025. https://doi.org/10.1186/s12864-016-3317-7
Kee N, Volakakis N, Kirkeby A, Dahl L, Storvall H, Nolbrant S, Lahti L, Bjorklund AK, Gillberg L, Joodmardi E, Sandberg R, Parmar M, Perlmann T (2017) Single-cell analysis reveals a close relationship between differentiating dopamine and subthalamic nucleus neuronal lineages. Cell Stem Cell 20(1):29–40. https://doi.org/10.1016/j.stem.2016.10.003
MacLean AL, Hong T, Nie Q (2018) Exploring intermediate cell states through the lens of single cells. Curr Opin Syst Biol 9:32–41. https://doi.org/10.1016/j.coisb.2018.02.009
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1):57–63. https://doi.org/10.1038/nrg2484
Rostom R, Svensson V, Teichmann SA, Kar G (2017) Computational approaches for interpreting scRNA-seq data. Febs Lett 591(15):2213–2225. https://doi.org/10.1002/1873-3468.12684
Tang FC, Barbacioru C, Wang YZ, Nordman E, Lee C, Xu NL, Wang XH, Bodeau J, Tuch BB, Siddiqui A, Lao KQ, Surani MA (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6(5):377-U386. https://doi.org/10.1038/nmeth.1315
Dulken BW, Leeman DS, Boutet SC, Hebestreit K, Brunet A (2017) Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep 18(3):777–790. https://doi.org/10.1016/j.celrep.2016.12.060
Zurauskiene J, Yau C (2016) pcaReduce: hierarchical clustering of single cell transcriptional profiles. BMC Bioinform. https://doi.org/10.1186/s12859-016-0984-y
Arendt D, Bertucci PY, Achim K, Musser JM (2019) Evolution of neuronal types and families. Curr Opin Neurobiol 56:144–152. https://doi.org/10.1016/j.conb.2019.01.022
Yang X, Gao L, Zhang S (2017) Comparative pan-cancer DNA methylation analysis reveals cancer common and specific patterns. Brief Bioinform 18(5):761–773. https://doi.org/10.1093/bib/bbw063
de la Fuente A (2010) From “differential expression” to “differential networking” - identification of dysfunctional regulatory networks in diseases. Trends Genet 26(7):326–333. https://doi.org/10.1016/j.tig.2010.05.001
West J, Bianconi G, Severini S, Teschendorff AE (2012) Differential network entropy reveals cancer system hallmarks. Sci Rep 2:802. https://doi.org/10.1038/srep00802
Lichtblau Y, Zimmermann K, Haldemann B, Lenze D, Hummel M, Leser U (2017) Comparative assessment of differential network analysis methods. Brief Bioinform 18(5):837–850. https://doi.org/10.1093/bib/bbw061
de Matos SR, Emmert-Streib F (2012) Bagging statistical network inference from large-scale gene expression data. PLoS ONE 7(3):e33624. https://doi.org/10.1371/journal.pone.0033624
Moerman T, Aibar Santos S, Bravo Gonzalez-Blas C, Simm J, Moreau Y, Aerts J, Aerts S (2019) GRNBoost2 and Arboreto: efficient and scalable inference of gene regulatory networks. Bioinformatics 35(12):2159–2161. https://doi.org/10.1093/bioinformatics/bty916
Liu H, Li P, Zhu M, Wang X, Lu J, Yu T (2016) Nonlinear network reconstruction from gene expression data using marginal dependencies measured by DCOL. PLoS ONE 11(7):e0158247. https://doi.org/10.1371/journal.pone.0158247
Stegle O, Teichmann SA, Marioni JC (2015) Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet 16(3):133–145. https://doi.org/10.1038/nrg3833
La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, Lonnerberg P, Ryge J, Barker RA, Arenas E, Linnarsson S (2016) Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167(2):566–580. https://doi.org/10.1016/j.cell.2016.09.027
Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, Bertagnolli D, Goldy J, Shapovalova N, Parry S, Lee C, Smith K, Bernard A, Madisen L, Sunkin SM, Hawrylycz M, Koch C, Zeng H (2016) Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19(2):335–346. https://doi.org/10.1038/nn.4216
Chen R, Wu X, Jiang L, Zhang Y (2017) Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep 18(13):3227–3241. https://doi.org/10.1016/j.celrep.2017.03.004
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Chasman D, Fotuhi Siahpirani A, Roy S (2016) Network-based approaches for analysis of complex biological systems. Curr Opin Biotechnol 39:157–166. https://doi.org/10.1016/j.copbio.2016.04.007
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW (2015) Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161(5):1187–1201. https://doi.org/10.1016/j.cell.2015.04.044
Nestorowa S, Hamey FK, Pijuan Sala B, Diamanti E, Shepherd M, Laurenti E, Wilson NK, Kent DG, Gottgens B (2016) A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128(8):e20-31. https://doi.org/10.1182/blood-2016-05-716480
Xie J, Yang F, Wang J, Karikomi M, Yin Y, Sun J, Wen T, Nie Q (2020) DNF: A differential network flow method to identify rewiring drivers for gene regulatory networks. Neurocomputing 410:202–210. https://doi.org/10.1016/j.neucom.2020.05.028
The Gene Ontology C (2019) The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res 47(D1):D330–D338. https://doi.org/10.1093/nar/gky1055
Bersanelli M, Mosca E, Remondini D, Castellani G, Milanesi L (2016) Network diffusion-based analysis of high-throughput data for the detection of differentially enriched modules. Sci Rep. https://doi.org/10.1038/srep34841
Picart-Armada S, Thompson WK, Buil A, Perera-Lluna A (2018) diffuStats: an R package to compute diffusion-based scores on biological networks. Bioinformatics 34(3):533–534. https://doi.org/10.1093/bioinformatics/btx632
Wang H, Li M, Wang J, Pan Y (2011) A new method for identifying essential proteins based on edge clustering coefficient. In: Bioinformatics research and applications. Berlin Heidelberg, Springer. pp 87–98
Koschützki D, Schreiber F (2008) Centrality analysis methods for biological networks and their application to gene regulatory networks. Gene Regul Syst Biol. https://doi.org/10.4137/grsb.s702
Odibat O, Reddy CK (2012) Ranking differential hubs in gene co-expression networks. J Bioinf Comput Biol. https://doi.org/10.1142/S0219720012400021
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
BarabÁSi A-L, Bonabeau E (2003) Scale-free networks. Sci Am 288(5):60–69
Canul-Tec JC, Assal R, Cirri E, Legrand P, Brier S, Chamot-Rooke J, Reyes N (2017) Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature 544(7651):446–451. https://doi.org/10.1038/nature22064
Hatanaka T, Huang W, Wang H, Sugawara M, Prasad PD, Leibach FH, Ganapathy V (2000) Primary structure, functional characteristics and tissue expression pattern of human, a subtype of amino acid transport system. Biochimica et Biophysica Acta (BBA) 1467(1):1–6. https://doi.org/10.1016/S0005-2736(00)00252-2
Hur EM, Zhou FQ (2010) GSK3 signalling in neural development. Nat Rev Neurosci 11(8):539–551. https://doi.org/10.1038/nrn2870
Rammensee S, Kang MS, Georgiou K, Kumar S, Schaffer DV (2018) Dynamics of mechanosensitive neural stem cell differentiation (vol 35, pg 497, 2017). Stem Cells 36(3):467–469. https://doi.org/10.1002/stem.2489
Perlini LE, Szczurkowska J, Ballif BA, Piccini A, Sacchetti S, Giovedi S, Benfenati F, Cancedda L (2015) Synapsin III acts downstream of semaphorin 3A/CDK5 signaling to regulate radial migration and orientation of pyramidal neurons in vivo. Cell Reports 11(2):234–248. https://doi.org/10.1016/j.celrep.2015.03.022
Whalley K (2009) NEURODEGENERATIVE DISEASE APP: what’s on the inside matters. Nat Rev Neurosci 10(12):836–836. https://doi.org/10.1038/nrn2760
Inestrosa NC, Varela-Nallar L (2015) Wnt signalling in neuronal differentiation and development. Cell Tissue Res 359(1):215–223. https://doi.org/10.1007/s00441-014-1996-4
Kuwajima T, Soares CA, Sitko AA, Lefebvre V, Mason C (2017) SoxC transcription factors promote contralateral retinal ganglion cell differentiation and axon guidance in the mouse visual system. Neuron 93(5):1110. https://doi.org/10.1016/j.neuron.2017.01.029
Frei JA, Stoeckli ET (2017) SynCAMs—from axon guidance to neurodevelopmental disorders. Mol Cell Neurosci 81:41–48. https://doi.org/10.1016/j.mcn.2016.08.012
Fuellen G (2011) Evolution of gene regulation–on the road towards computational inferences. Brief Bioinform 12(2):122–131. https://doi.org/10.1093/bib/bbq060
Funding
This work was partially supported by the National Natural Science Foundation of China [No.61873156], the Shanghai Municipal Science and Technology Major Project [No.2018SHZDZX01], Basic Research Program of Shanghai (20JC1412200), Key Laboratory of Computational Neuroscience and Brain-Inspired Intelligence (LCNBI) and ZJLab.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xie, J., Yin, Y., Yang, F. et al. Differential Network Analysis Reveals Regulatory Patterns in Neural Stem Cell Fate Decision. Interdiscip Sci Comput Life Sci 13, 91–102 (2021). https://doi.org/10.1007/s12539-020-00415-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-020-00415-2