Abstract
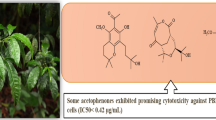
In the present investigation, QSAR analysis was performed on a data set consist of structurally diverse compounds in order to investigate the role of its structural features on their Photosynthetic Electron Transport Inhibitors. The herbicidal activity co-related with certain topological and hydrophobicity based descriptors, 3D descriptors dependent steric, electrostatic and hydrophobic. The best 2D QSAR model was selected, having correlation coefficient r2 = 0.8544 and cross validated squared correlation coefficient q2 = 0.7139 with external predictive ability of pred_r2 = 0.7753 was developed. The results obtained in this study indicate that hydroxy and nitro groups, as expressed by the SsOHcount, SddsN (nitro) count, is the most relevant molecular property determining efficiency of photosynthetic inhibitory. Molecular field analysis was used to construct the best k-nearest neighbor (kNN-MFA)-based 3DQSAR model using SA-PLS method, showing good correlative and predictive capabilities in terms of q2 = 0.7694 and pred_r2 = 0.7381. The influences of steric, electrostatic and hydrophobic field effects generated by the contribution plots are discussed. The pharmacophore model includes three features viz. hydrogen bond donor, hydrogen bond acceptor, and one aromatic feature was developed. The developed model was found to be predictive and can be used to design potent Photosynthetic Electron Transport activities prior to their synthesis for further lead modification. The results obtained suggest that the 3-nitro-2, 4, 6-trihydroxybenzamide analogues represent promising candidates for the development of new active principles targeting photosynthesis to be used as herbicides.
Similar content being viewed by others
References
Ajmani S, Agrawal A, Kulkarni SA. A comprehensive structure-activity analysis of protein kinase B-alpha (Akt1) inhibitors. J Mol Graph Model 2010; 28: 683–694.
Ajmani S, Jhadav K, Kulkarni SA. Three-dimensional QSAR using the k-nearest neighbor method and its interpretation. J Chem Inf Mod 2006; 46: 24–31.
Baumann K. An alignment-independent versatile structure descriptor for QSAR and QSPR based on the distribution of molecular features. J Chem Inf Comput Sci 2000; 42: 26–35.
Bhatia MS, Pakhare KD, Choudhari PB, Jadhav S D, Dhavale RP, Bhatia NM (2012) Pharmacophore modeling and 3D QSAR studies of aryl amine derivatives as potential lumazine synthase inhibitors. Arab J Chem https://doi.org/10.1016/j.arabjc.2012.05.008.
Bhatiya R, Vaidya A, Kashaw SK, Jain AK, Agrawal AK (2011) QSAR analysis of furanone derivatives as potential COX-2 inhibitors: kNN MFA approach. J Saud Chem Soc doi: https://doi.org/10.1016/j.jscs.2011.12.002.
Choudhari P, Bhatia M (2012) 3D QSAR, pharmacophore indentification studies on series of 1-(2-ethoxyethyl)-1Hpyrazolo [4, 3-d] pyrimidines as phosphodiesterase V inhibitors. J Saudi Chem Soc https://doi.org/10.1016/j.jscs.2012.02.008.
Clark M, Cramer RD III, Van ON. Validation of the general purpose tripos 5.2 force field. J Comput Chem 1989; 10: 982–1012.
Cramer RD, Patterson DE, Bunce JD (1988) Comparative Molecular Field Analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110, 5959–5967.
Darlington R.B. Regression and Linear Models, McGraw-Hill, New York, 1990.
Devine MD, Shukla A. Altered target sites as a mechanism of herbicide resistance. Crop Protection 2009; 19: 881–889.
Ghosh P, Bagchi MC. QSAR modeling for quinoxaline derivatives using genetic algorithm and simulated annealing based feature selection. Curr Med Chem 2009; 16: 4032–4048.
Gasteiger J, Marsili M. Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron 1980; 36: 3219–3228.
Golbraikh A, Tropsha A. Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. J Comput Aided Mol Des 2002; 16: 357–369.
Halgren TA. Merck molecular force field. III. Molecular geometries and vibrational frequencies for MMFF94. J Comput Chem 1996; 17: 553–586.
Holland JH. Genetic algorithms. Sci Am 1992; 267: 66–72.
Han XF, Liu YX, Liu Y, Lai LH, Huang RQ, Wang QM. Binding model and 3DQSAR of 3-(2-chloropyrid- 5-ylmethylamino)-2-cyanoacrylates as PSII electron transport inhibitor. Chin J Chem 2007; 25: 1135–1138.
Honda I, Yoneyama K, Konnai M, Takahashi N, Yoshida S. Structure activity relationship 3-Nitro-2, 4, 6-trihydroxybenzamide and Thiobenzamide Derivatives in Photosynthetic Electron Transport inhibition. Agr Biolog Chem 1990; 54(5): 1227–1233.
Hansch C, Leo A. Exploring QSAR: Fundamentals and Applications in Chemistry and Biology, American Chemical Society, Washington, 1995.
Heap I. International survey of herbicide resistant weeds, http://www.weedscience.org, accessed Dec 28, 2004.
Kubinyi H., QSAR Hansch Analysis and Related Approaches, VCH, New York, 1993.
Kubinyi H. QSAR and 3DQSAR in drug design part 1: methodology. Drug Discov Today 1997; 2: 457–467.
Karacan MS, Yakan Ç, Yakan M, Karacan N, Zharmukhamedov SK, Shitov A, Los D A, Klimov V V, Allakhverdiev S I. Quantitative structure-activity relationship analysis of perfluoroiso-propyldinitrobenzene derivatives known as photosystem II electron transfer inhibitors. Biochimica et Biophysica Acta 2012; 1817: 1229–1236.
Lein W, Bornke F, Reindl A, Ehrhardt T, Stitt M, Sonnewald U. Target-based discovery of novel herbicides. Curr Opin Plant Biol 2004; 7: 219–225.
Mandloi D, Joshi S, Khadikar PV, Khosla N. QSAR study on the antibacterial activity of some sulfadrugs building blockers of Mannich bases. Bioorg Med Chem Lett 2005; 15: 405–411.
Oettmeier W. Herbicide resistance and super sensitivity in photosystem II. Cellular and Molecular Life Sciences 1999; 55: 1255–1277.
Sharma PC, Sharma SV, Singh D, Suresh B. QSAR Studies on 3-Nitro-2, 4, 6-trihydroxy Benzamide derivatives as Photosynthetic Electron Transport inhibitors. Asian J Chem 2007; 19 (17): 5105–5112.
Shimizu R, Iwamura H, Fujita T. Quantitative structure activity relationships of photosystem-II inhibitory anilides and triazines-topological aspects of their binding to the active-site. J Agr Food Chem 1988: 36: 1276–1283.
Sahu NK, Shahi S, Sharma MC, Kohli DV, QSAR studies on imidazopyridazine derivatives as PfPK7 inhibitors. Mol Simul 2011; 37(9): 752–765.
Teixeira R R, Pinheiro PF, Barbosa L C A, Carneiro JWM, Forlanic G. QSAR modeling of photosynthesisinhibiting nostoclide derivatives. Pest Manag Sci 2010; 66: 196–202.
Vicentini BC, Guccione S, Giurato L, Ciaccio R, Mares D, Forlani G. Pyrazole Derivatives as Photosynthetic Electron Transport Inhibitors: New Leads and Structure-Activity Relationship. J Agr Food Chem 2005; 53: 3848–3855.
VLife MDS 3.5 (2008) Molecular design suite. Vlife Sciences Technologies Pvt. Ltd., Pune. www.vlifesciences.com.
Wold S (1995) in QSAR-Chemometric Methods in Molecular Design, Vol. 2 (Ed: H. van de Waterbeemd), Wiley-VCH, Weinheim, Germany, 1995, pp. 195–218.
Wakabayashi K, Boger P. Target sites for herbicides: entering the 21st century. Pest Manag Scien 2002; 58: 1149–1154.
Zheng W, Tropsha A. Novel variable selection quantitative structure-property relationship approach based on the k-nearest neighbor principle. J Chem Inf Comput Sci 2000; 40: 185–194.
Zakarya D, Larfaoui EM, Boulaamail A, Toilabi M, Lakhlifi T. QSARs for a series of inhibitory anilides. Chemosphere 1988; 36: 2809–2818.
Acknowledgements
The author would like to thank VLife Sciences Technologies Pvt. Ltd. Pune for providing trial version software facility.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharma, M.C. Identification of 3-Nitro-2, 4, 6-Trihydroxybenzamide Derivatives as Photosynthetic Electron Transport Inhibitors by QSAR and Pharmacophore Studies. Interdiscip Sci Comput Life Sci 8, 109–121 (2016). https://doi.org/10.1007/s12539-015-0019-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-015-0019-9