Abstract
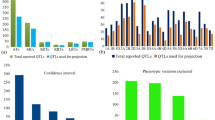
In wheat, meta-QTLs (MQTLs), ortho-MQTLs, and candidate genes (CGs) were identified for nitrogen use efficiency and root system architecture. For this purpose, 1788 QTLs were available from 24 studies published during 2006–2020. Of these, 1098 QTLs were projected onto the consensus map resulting in 118 MQTLs. The average confidence interval (CI) of MQTLs was reduced up to 8.56 folds in comparison to the average CI of QTLs. Of the 118 MQTLs, 112 were anchored to the physical map of the wheat reference genome. The physical interval of MQTLs ranged from 0.02 to 666.18 Mb with a mean of 94.36 Mb. Eighty-eight of these 112 MQTLs were verified by marker-trait associations (MTAs) identified in published genome-wide association studies (GWAS); the MQTLs that were verified using GWAS also included 9 most robust MQTLs, which are particularly useful for breeders; we call them ‘Breeder’s QTLs’. Some selected wheat MQTLs were further utilized for the identification of ortho-MQTLs for wheat and maize; 9 such ortho-MQTLs were available. As many as 1991 candidate genes (CGs) were also detected, which included 930 CGs with an expression level of > 2 transcripts per million in relevant organs/tissues. Among the CGs, 97 CGs with functions previously reported as important for the traits under study were selected. Based on homology analysis and expression patterns, 49 orthologues of 35 rice genes were also identified in MQTL regions. The results of the present study may prove useful for the improvement of selection strategy for yield potential, stability, and performance under N-limiting conditions.








Similar content being viewed by others
Availability of data and material
Data generated or analysed during this study are included in this published article (and its Supplementary Material).
References
Aduragbemi A, Soriano JM (2021) Unravelling consensus genomic regions conferring leaf rust resistance in wheat via meta-QTL analysis. bioRxiv. doi:https://doi.org/10.1101/2021.05.11.443557
Ahn S, Anderson JA, Sorrells ME, Tanksley SD (1993) Homoeologous relationships of rice, wheat and maize chromosomes. Mol Gen Genet 241:483–490. https://doi.org/10.1007/BF00279889
Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20:2324–2326. https://doi.org/10.1093/bioinformatics/bth230
Balyan HS, Gahlaut V, Kumar A, Jaiswal V, Dhariwal R, Tyagi S, Agarwal P, Kumari S, Gupta PK (2016) Nitrogen and phosphorus use efficiencies in wheat: physiology, phenotyping, genetics, and breeding. Plant Breed Rev 40:167–234. https://doi.org/10.1002/9781119279723.ch4
Beatty PH, Carroll RT, Shrawat AK, Guevara D, Good AG (2013) Physiological analysis of nitrogen-efficient rice overexpressing alanine aminotransferase under different N regimes. Botany 91:866–883
Bennetzen JL, Chen M (2008) Grass genomic synteny illuminates plant genome function and evolution. Rice 1:109–118. https://doi.org/10.1007/s12284-008-9015-6
Bi YM, Kant S, Clark J, Gidda S, Ming F, Xu J, Rochon A, Shelp BJ, Hao L, Zhao R, Mullen RT (2009) Increased nitrogen-use efficiency in transgenic rice plants over-expressing a nitrogen-responsive early nodulin gene identified from rice expression profiling. Plant Cell Environ 32:1749–1760
Brasier K, Ward B, Smith J, Seago J, Oakes J, Balota M, Davis P, Fountain M, Brown-Guedira G, Sneller C, Thomason W (2020) Identification of quantitative trait loci associated with nitrogen use efficiency in winter wheat. PLoS ONE 15:e0228775. https://doi.org/10.1371/journal.pone.0228775
Chiasson DM, Loughlin PC, Mazurkiewicz D, Mohammadidehcheshmeh M, Fedorova EE, Okamoto M, McLean E, Glass AD, Smith SE, Bisseling T, Tyerman SD (2014) Soybean SAT1 (Symbiotic Ammonium Transporter 1) encodes a bHLH transcription factor involved in nodule growth and NH4+ transport. Proc Natl Acad Sci 111:4814–4819. https://doi.org/10.1073/pnas.1312801111
Cirilo AG, Dardanelli J, Balzarini M, Andrade FH, Cantarero M, Luque S, Pedrol HM (2009) Morpho-physiological traits associated with maize crop adaptations to environments differing in nitrogen availability. Field Crops Res 113:116–124. https://doi.org/10.1016/j.fcr.2009.04.011
Cobb JN, Biswas PS, Platten JD (2019) Back to the future: revisiting MAS as a tool for modern plant breeding. Theor Appl Genet 132:647–667. https://doi.org/10.1007/s00122-018-3266-4
Coello P, Hey SJ, Halford NG (2011) The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J Exp Bot 62:883–893. https://doi.org/10.1093/jxb/erq331
Collard BCY, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 363:557–572
Dalton DA, Boniface C, Turner Z, Lindahl A, Kim HJ, Jelinek L, Govindarajulu M, Finger RE, Taylor CG (2009) Physiological roles of glutathione S-transferases in soybean root nodules. Plant Physiol 150:521–530
Darvasi A, Soller M (1997) A simple method to calculate resolving power and confidence interval of QTL map location. Behav Genet 27:125–132. https://doi.org/10.1023/a:1025685324830
Darzi-Ramandi H, Shariati J V, Tavakol E, Najafi-Zarini H, Bilgrami SS, Razavi K (2017) Detection of consensus genomic regions associated with root architecture of bread wheat on groups 2 and 3 chromosomes using QTL meta–analysis. Aust J Crop Sci 777–785.
Do THT, Martinoia E, LeeY, (2018) Functions of ABC transporters in plant growth and development. Curr Opin Plant Biol 41:32–38
Dong NQ, Sun Y, Guo T, Shi CL, Zhang YM, Kan Y, Xiang YH, Zhang H, Yang YB, Li YC, Zhao HY (2020) UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat Commun 11:1–16
Endelman JB, Plomion C (2014) LPmerge: an R package for merging genetic maps by linear programming. Bioinformatics 30:1623–1624. https://doi.org/10.1093/bioinformatics/btu091
Esposito S, Guerriero G, Vona V, Di Martino RV, Carfagna S, Rigano C (2005) Glutamate synthase activities and protein changes in relation to nitrogen nutrition in barley: the dependence on different plastidic glucose-6P dehydrogenase isoforms. J Exp Bot 56:55–64
Fan X, Cui F, Ji J, Zhang W, Zhao X, Liu J, Meng D, Tong Y, Wang T, Li J (2019) Dissection of pleiotropic QTL regions controlling wheat spike characteristics under different nitrogen treatments using traditional and conditional QTL mapping. Front Plant Sci 10:187. https://doi.org/10.3389/fpls.2019.00187
Forde BG (2014) Nitrogen signaling pathways shaping root system architecture: an update. Curr Opin Plant Biol 21:30–36
Garnett T, Conn V, Kaiser BN (2009) Root based approaches to improving nitrogen use efficiency in plants. Plant Cell Environ 32:1272–1283. https://doi.org/10.1111/j.1365-3040.2009.02011.x
Gaut BS (2002) Evolutionary dynamics of grass genomes. New Phytol 154:15–28
Goffinet B, Gerber S (2000) Quantitative trait loci: a meta-analysis. Genetics 155:463–473
Good AG, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9:597–605. https://doi.org/10.1016/j.tplants.2004.10.008
Good AG, Beatty PH (2011) Biotechnological approaches to improving nitrogen use efficiency in plants: alanine aminotransferase as a case study. In: Malcolm JH, Peter B (ed) The molecular and physiological basis of nutrient use efficiency in crops. Wiley, New York, pp 165–191. doi:https://doi.org/10.1002/9780470960707
Greef JM (1994) Productivity of maize (Zea mays L.) in relation to morphological and physiological characteristics under varying amounts of nitrogen supply. J Agron Crop Sci 172:317–326
Guo B, Sleper DA, Lu P, Shannon JG, Nguyen HT, Arelli PR (2006) QTLs associated with resistance to soybean cyst nematode in soybean: meta-analysis of QTL locations. Crop Sci 46:595–602
Guo J, Chen L, Li Y, Shi Y, Song Y, Zhang D, Li Y, Wang T, Yang D, Li C (2018) Meta-QTL analysis and identification of candidate genes related to root traits in maize. Euphytica 214:223. https://doi.org/10.1007/s10681-018-2283-3
Han M, Wong J, Su T, Beatty PH, Good AG (2016) Identification of nitrogen use efficiency genes in barley: searching for QTLs controlling complex physiological traits. Front Plant Sci 7:1587
Han G, Lu C, Guo J, Qiao Z, Sui N, Qiu N, Wang B (2020) C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front Plant Sci 11:115. https://doi.org/10.3389/fpls.2020.00115
Hawkesford MJ (2012) Improving nutrient use efficiency in crops. eLS. doi:https://doi.org/10.1002/9780470015902.a0023734
He X, Qu B, Li W, Zhao X, Teng W, Ma W, Ren Y, Li B, Li Z, Tong Y (2015) The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol 169:1991–2005
Hirel B, Le Gouis J, Ney B, Gallais A (2007) The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58:2369–2387
Jan I, Saripalli G, Kumar K, Kumar A, Singh R, Batra R, Sharma PK, Balyan HS, Gupta PK (2021) Meta-QTL analysis for stripe rust resistance in wheat. doi:https://doi.org/10.21203/rs.3.rs-380807/v1
Jun XU, Wang XY, Guo WZ (2015) The cytochrome P450 superfamily: key players in plant development and defense. J Integr Agr 14:1673–1686
Karunarathne SD, Han Y, Zhang XQ, Li C (2020) Advances in understanding the molecular mechanisms and potential genetic improvement for nitrogen use efficiency in barley. Agronomy 10:662. https://doi.org/10.3390/agronomy10050662
Khahani B, Tavakol E, Shariati V, Fornara F (2020) Genome wide screening and comparative genome analysis for Meta-QTLs, ortho-MQTLs and candidate genes controlling yield and yield-related traits in rice. BMC Genom 21:294. https://doi.org/10.1186/s12864-020-6702-1
Khahani B, Tavakol E, Shariati V, Rossini L (2021) Meta-QTL and ortho-MQTL analyses identified genomic regions controlling rice yield, yield-related traits and root architecture under water deficit conditions. Sci Rep 11:6942. https://doi.org/10.1038/s41598-021-86259-2
Kumar S, Mohan A, Balyan HS, Gupta PK (2009) Orthology between genomes of Brachypodium, wheat and rice. BMC Res Notes 2:1–9
Kumar IS, Nadarajah K (2020) A meta-analysis of quantitative trait loci associated with multiple disease resistance in rice (Oryza sativa L.). Plants 9:1491. doi:https://doi.org/10.3390/plants9111491
Kumari S, Raghuram N (2020) Protein Phosphatases in N Response and NUE in Crops. In: Pandey G (ed) Protein phosphatases and stress management in plants. Springer, Switzerland, pp 233–244
Kumari S, Sharma N, Raghuram N (2021) Meta-analysis of yield-related and N-responsive genes reveals chromosomal hotspots, key processes and candidate genes for nitrogen-use efficiency in rice. Front Plant Sci 12:627955
Lahners K, Kramer V, Back E, Privalle L, Rothstein S (1988) Molecular cloning of complementary DNA encoding maize nitrite reductase: molecular analysis and nitrate induction. Plant Physiol 88:741–746
Li P, Chen F, Cai H, Liu J, Pan Q, Liu Z, Gu R, Mi G, Zhang F, Yuan L (2015) A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J Exp Bot 66:3175–3188. https://doi.org/10.1093/jxb/erv127
Li X, Zeng R, Liao H (2016) Improving crop nutrient efficiency through root architecture modifications. J Integr Plant Biol 58:193–202
Li M, Xu J, Gao Z, Tian H, Gao Y, Kariman K (2020) Genetically modified crops are superior in their nitrogen use efficiency: a meta-analysis of three major cereals. Sci Rep 10:1–9
Liu R, Zhang H, Zhao P, Zhang Z, Liang W, Tian Z, Zheng Y (2012) Mining of candidate maize genes for nitrogen use efficiency by integrating gene expression and QTL data. Plant Mol Biol Rep 30:297–308. https://doi.org/10.1007/s11105-011-0346-x
López-Bucio J, Cruz-Ramırez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6:280–287
Marone D, Russo MA, Laidò G, De Vita P, Papa R, Blanco A, Gadaleta A, Rubiales D, Mastrangelo AM (2013) Genetic basis of qualitative and quantitative resistance to powdery mildew in wheat: from consensus regions to candidate genes. BMC Genom 14:562. https://doi.org/10.1186/1471-2164-14-562
Marowa P, Ding A, Kong Y (2016) Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep 35:949–965
Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot 105:1141–1157
Meijón M, Satbhai SB, Tsuchimatsu T, Busch W (2014) Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat Genet 46:77–81. https://doi.org/10.1038/ng.2824
Meister R, Rajani MS, Schachtman RD, DP, (2014) Challenges of modifying root traits in crops for agriculture. Trends Plant Sci 19:779–788
Minic Z (2008) Physiological roles of plant glycoside hydrolases. Planta 227:723–740
Misztal I (2006) Challenges of application of marker assisted selection: a review. Anim Sci Pap Rep 24:5–10
Moll RH, Kamprath EJ, Jackson WA (1982) Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization1. Agron J 74:562. https://doi.org/10.2134/agronj1982.00021962007400030037x
Mosleth EF, Wan Y, Lysenko A, Chope GA, Penson SP, Shewry PR, Hawkesford MJ (2015) A novel approach to identify genes that determine grain protein deviation in cereals. Plant Biotechnol J 13:625–635
Oaks A (1994) Primary nitrogen assimilation in higher plants and its regulation. Can J Bot 72:739–750
de Oliveira Y, Sosnowski O, Charcosset A, Joets J (2014) BioMercator 4: a complete framework to integrate QTL, meta-QTL, and genome annotation. In: European Conference on Computational Biology 2014 Sep 2014, Strasbourg, France
Qu B, He X, Wang J, Zhao Y, Teng W, Shao A, Zhao X, Ma W, Wang J, Li B, Li Z (2015) A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol 167:411–423
Quraishi UM, Abrouk M, Bolot S, Pont C, Throude M, Guilhot N, Confolent C, Bortolini F, Praud S, Murigneux A, Charmet G (2009) Genomics in cereals: from genome-wide conserved orthologous set (COS) sequences to candidate genes for trait dissection. Funct Integr Genom 9:473–484
Quraishi UM, Abrouk M, Murat F, Pont C, Foucrier S, Desmaizieres G, Confolent C, Riviere N, Charmet G, Paux E, Murigneux A (2011) Cross-genome map based dissection of a nitrogen use efficiency ortho-metaQTL in bread wheat unravels concerted cereal genome evolution. Plant J 65:745–756. https://doi.org/10.1111/j.1365-313X.2010.04461.x
Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, Van Ex F, Pasha A, Khedikar Y (2018) The transcriptional landscape of polyploid wheat. Science. https://doi.org/10.1126/science.aar6089
Raun WR, Johnson GV (1999) Improving nitrogen use efficiency for cereal production. Agronomy 91:357–363
Saini DK, Srivastava P, Pal N, Gupta PK (2021) Meta-QTLs, ortho-metaQTLs and candidate genes for grain yield and associated traits in wheat (Triticum aestivum L.). doi:https://doi.org/10.21203/rs.3.rs-430452/v1
Salarpour M, Pakniyat H, Abdolshahi R, Heidari B, Razi H, Afzali R (2020) Mapping QTL for agronomic and root traits in the Kukri/RAC875 wheat (Triticum aestivum L.) population under drought stress conditions. Euphytica 216:1–19
Singh K, Batra R, Sharma S, Saripalli G, Gautam T, Singh R, Pal S, Malik P, Kumar M, Jan I, Singh S (2021) WheatQTLdb: a QTL database for wheat. Mol Genet Genom. https://doi.org/10.1007/s00438-021-01796-9
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114. https://doi.org/10.1007/s00122-004-1740-7
Soriano JM, Alvaro F (2019) Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci Rep 9:10537. https://doi.org/10.1038/s41598-019-47038-2
Sosnowski O, Charcosset A, Joets J (2012) BioMercator V3: an upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 28:2082–2083. https://doi.org/10.1093/bioinformatics/bts313
Sun JJ, Guo Y, Zhang GZ, Gao MG, Zhang GH, Kong FM, Zhao Y, Li SS (2013) QTL mapping for seedling traits under different nitrogen forms in wheat. Euphytica 191:317–331. https://doi.org/10.1007/s10681-012-0834-6
Sun Y, Hu Z, Wang X, Shen X, Hu S, Yan Y, Kant S, Xu G, Xue Y, Sun S (2021) Overexpression of OsPHR3 improves growth traits and facilitates nitrogen use efficiency under low phosphate condition. Plant Physiol Biochem 166:712–722
Takahashi M, Sasaki Y, Ida S, Morikawa H (2001) Nitrite reductase gene enrichment improves assimilation of NO2 in Arabidopsis. Plant Physiol 126:731–741
Tanaka S, Ida S, Irifune K, Oeda K, Morikawa H (1994) Nucleotide sequence of a gene for nitrite reductase from Arabidopsis thaliana. DNA Seq 5:57–61
Tiong J, Sharma N, Sampath R, MacKenzie N, Watanabe S, Metot C, Lu Z, Skinner W, Lu Y, Kridl J, Baumann U (2021) Improving nitrogen use efficiency through overexpression of alanine aminotransferase in rice, wheat, and barley. Front Plant Sci 12:29
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314:1298–1301
Vanoni MA, Curti B (1999) Glutamate synthase: a complex iron-sulfur flavoprotein. Cell Mol Life Sci 55:617–638
Venske E, Dos Santos RS, Farias DD, Rother V, da Maia LC, Pegoraro C, Costa de Oliveira A (2019) Meta-analysis of the QTLome of fusarium head blight resistance in bread wheat: Refining the current puzzle. Front Plant Sci 10:727. https://doi.org/10.3389/fpls.2019.00727
Veyrieras J-B, Goffinet B, Charcosset A (2007) MetaQTL: a package of new computational methods for the meta-analysis of QTL mapping experiments. BMC Bioinform 8:49. https://doi.org/10.1186/1471-2105-8-49
Vidal EA, Alvarez JM, Araus V, Riveras E, Brooks MD, Krouk G, Ruffel S, Lejay L, Crawford NM, Coruzzi GM, Gutiérrez RA (2020) Nitrate in 2020: thirty years from transport to signaling networks. Plant Cell 32:2094–2119
Wagner GP, Kin K, Lynch VJ (2013) A model based criterion for gene expression calls using RNA-seq data. Theory Biosci 132:159–164. https://doi.org/10.1007/s12064-013-0178-3
Xiang J, Qian K, Zhang Y, Chew J, Liang J, Zhu J, Zhang Y, Fan X (2021) OsLSD1.1 is involved in the photosystem II reaction and affects nitrogen allocation in rice. Plant Physiol Biochem. https://doi.org/10.1016/j.plaphy.2021.06.004
Xu J, Wang X, Guo W (2015) The cytochrome P450 superfamily: Key players in plant development and defense. J Integr Agric 14:1673–1686. https://doi.org/10.1016/S2095-3119(14)60980-1
Yamaya T (2011) Disruption of a novel NADH-glutamate synthase2 gene caused marked reduction in spikelet number of rice. Front Plant Sci 2:57
Yang X, Xia X, Zeng Y, Nong B, Zhang Z, Wu Y, Tian Q, Zeng W, Gao J, Zhou W, Liang H (2020) Genome-wide identification of the peptide transporter family in rice and analysis of the PTR expression modulation in two near-isogenic lines with different nitrogen use efficiency. BMC Plant Biol 20:193. https://doi.org/10.1186/s12870-020-02419-y
Yang Y, Amo A, Wei D, Chai Y, Zheng J, Qiao P, Cui C, Lu S, Chen L, Hu YG (2021) Large-scale integration of meta-QTL and genome-wide association study discovers the genomic regions and candidate genes for yield and yield-related traits in bread wheat. Theor Appl Genet 134:1–27
Yu J, Xuan W, Tian Y, Fan L, Sun J, Tang W, Chen G, Wang B, Liu Y, Wu W, Liu X (2021) Enhanced OsNLP4-OsNiR cascade confers nitrogen use efficiency by promoting tiller number in rice. Plant Biotechnol J 19:167–176
Zhang Y, Yu C, Lin J, Liu J, Liu B, Wang J, Huang A, Li H, Zhao T (2017) OsMPH1 regulates plant height and improves grain yield in rice. PLoS ONE 12:e0180825. https://doi.org/10.1371/journal.pone.0180825
Zhang N, Zhang X, Song L, Su Q, Zhang S, Liu J, Zhang W, Fu X, Zhao M, Sun L, Ji J (2020a) Identification and validation of the superior alleles for wheat kernel traits detected by genome-wide association study under different nitrogen environments. Euphytica 216:1–15
Zhang Z, Gao S, Chu C (2020b) Improvement of nutrient use efficiency in rice: current toolbox and future perspectives. Theor Appl Genet 133:1365–1384
Zhao X, Peng Y, Zhang J, Fang P, Wu B (2018) Identification of QTLs and meta-QTLs for seven agronomic traits in multiple maize populations under well-watered and water-stressed conditions. Crop Sci 58:507–520
Acknowledgements
The Department of Science and Technology (DST) in New Delhi, India, provided DKS with an INSPIRE fellowship, while the Head, Department of Plant Breeding and Genetics, Punjab Agricultural University in Ludhiana, India, provided the essential facilities.
Author information
Authors and Affiliations
Contributions
PKG and PS conceived and planned the study. DKS, YC, NP and AC collected the literature and tabulated the data for meta-QTL analysis. DKS conducted the analysis. DKS and YC interpreted the results and wrote the manuscript. PKG and PS edited and finalized the manuscript with the help of DKS.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saini, D.K., Chopra, Y., Pal, N. et al. Meta-QTLs, ortho-MQTLs and candidate genes for nitrogen use efficiency and root system architecture in bread wheat (Triticum aestivum L.). Physiol Mol Biol Plants 27, 2245–2267 (2021). https://doi.org/10.1007/s12298-021-01085-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-01085-0