Abstract
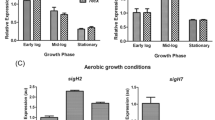
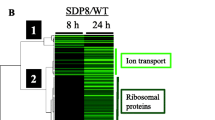
Saccharomyces cerevisiae Hugl is a small protein of unknown function that is highly inducible following replication stress and DNA damage. Its deletion suppresses the lethality of deletion of checkpoint kinase Mecl. Although DNA damage responses were largely normal in the HUG1 deletion mutant, we found enhanced resistance towards heat in logarithmic phase. In response to simultaneous carbon and replication stress, overall growth delay and less pseudohyphal filament formation were evident. These novel phenotypes are shared with deletion mutants of the negative regulators of ribonucleotide reductase, Difl and Smll. Microarray analysis showed the influence of Hugl on the expression of a large number of transcripts, including stress-related transcripts. Elevated dNTP levels in hugl Δ cells may result in a stress response reflected by the observed phenotypes and transcript profiles. However, in contrast to a deletion of structurally related Difl, Rnr2-Rnr4 subcellular localization is not grossly altered in a Hugl deletion mutant. Thus, although Hugl appears to be derived from the Rnr2-Rnr4 binding region of Difl, its mechanism of action must be independent of determining the localization of Rnr2-Rnr4.
Similar content being viewed by others
References
Basrai, M.A., V.E. Velculescu, K.W. Kinzler, and P. Hieter. 1999. NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 7041–7049.
Benton, M.G., N.R. Glasser, and S.P. Palecek. 2007. The utilization of a Saccharomyces cerevisiae HUG1P-GFP promoter-reporter construct for the selective detection of DNA damage. Mutat. Res. 633, 21–34.
Carroll, A.S., A.C. Bishop, J.L. DeRisi, K.M. Shokat, and E.K. O’shea. 2001. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc. Natl. Acad. Sci. USA 98, 12578–12583.
Chabes, A., B. Georgieva, V. Domkin, X. Zhao, R. Rothstein, and L. Thelander. 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 112, 391–401.
Friedberg, E.C., G.C. Walker, W. Siede, R.D. Wood, R.A. Schultz, and T. Ellenberger. 2006. DNA Repair and Mutagenesis, 2nd (ed.). American Society of Microbiology Press, Washington, D.C., USA.
Gasch, A.P. and M. Werner-Washburne. 2002. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2, 181–192.
Huang, D., H. Friesen, and B. Andrews. 2007. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol. Microbiol. 66, 303–314.
Huang, M., Z. Zhou, and S.J. Elledge. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 94, 595–605.
Jiang, Y.W. and C.M. Kang. 2003. Induction of S. cerevisiae filamentous differentiation by slowed DNA synthesis involves Mec1, Rad53 and Swe1 checkpoint proteins. Mol. Biol. Cell. 14, 5116–5124.
Kastan, M.B. and J. Bartek. 2004. Cell-cycle checkpoints and cancer. Nature 432, 316–323.
Lazzaro, F., M. Giannattasio, F. Puddu, M. Granata, A. Pellicioli, P. Plevani, and M. Muzi-Falconi. 2009. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair (Amst). 8, 1055–1067.
Lee, Y.D. and S.J. Elledge. 2006. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 20, 334–344.
Lee, M.W., B.J. Kim, H.K. Choi, M.J. Ryu, S.B. Kim, K.M. Kang, E.J. Cho, H.D. Youn, W.K. Huh, and S.T. Kim. 2007. Global protein expression profiling of budding yeast in response to DNA damage. Yeast 24, 145–154.
Lee, Y.D., J. Wang, J. Stubbe, and S.J. Elledge. 2008. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol. Cell. 32, 70–80.
Lussier, M., A.M. White, J. Sheraton, T. di Paolo, J. Treadwell, S.B. Southard, C.I. Horenstein, and et al. 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147, 435–450.
Mizukami-Murata, S., H. Iwahashi, S. Kimura, K. Nojima, Y. Sakurai, T. Saitou, N. Fujii, and et al. 2010. Genome-wide expression changes in Saccharomyces cerevisiae in response to high-LET ionizing radiation. Appl. Biochem. Biotechnol. 162, 855–870.
Nestoras, K., A.H. Mohammed, A.S. Schreurs, O. Fleck, A.T. Watson, M. Poitelea, C. O’shea, and et al. 2010. Regulation of ribonucleotide reductase by Spd1 involves multiple mechanisms. Genes Dev. 24, 1145–1159.
Nyberg, K.A., R.J. Michelson, C.W. Putnam, and T.A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36, 617–656.
Pabla, R., V. Pawar, H. Zhang, and W. Siede. 2006. Characterization of checkpoint responses to DNA damage in Saccharomyces cerevisiae: basic protocols. Meth. Enzymol. 409, 101–117.
Ptacek, J., G. Devgan, G. Michaud, H. Zhu, X. Zhu, J. Fasolo, H. Guo, and et al. 2005. Global analysis of protein phosphorylation in yeast. Nature 438, 679–684.
Rothstein, R. 1989. Targeting, disruption, replacement, and allelic rescue: integrative DNA transformation in yeast. Meth. Enzymol. 194, 281–301.
Wu, X. and M. Huang. 2008. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol. Cell. Biol. 28, 7156–7167.
Yao, R., Z. Zhang, X. An, B. Bucci, D.L. Perlstein, J. Stubbe, and M. Huang. 2003. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Genetics 100, 6628–6633.
Zaim, J., E. Speina, and A.M. Kierzek. 2005. Identification of new genes regulated by the Crt1 transcription factor, an effector of the DNA damage checkpoint pathway in Saccharomyces cerevisiae. J. Biol. Chem. 280, 28–37.
Zhao, X., A. Chabes, V. Domkin, L. Thelander, and R. Rothstein. 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20, 3544–3553.
Zhao, X., B. Georgieva, A. Chabes, V. Domkin, J.H. Ippel, J. Schleucher, S. Wijmenga, L. Thelander, and R. Rothstein. 2000. Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol. Cell. Biol. 20, 9076–9083.
Zhao, X. and R. Rothstein. 2002. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA 99, 3746–3751.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at http://www.springer.com/content/120956
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kim, E., Siede, W. Phenotypes associated with Saccharomyces cerevisiae Hug1 protein, a putative negative regulator of dNTP Levels, reveal similarities and differences with sequence-related dif1. J Microbiol. 49, 78–85 (2011). https://doi.org/10.1007/s12275-011-0200-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-011-0200-8