Abstract
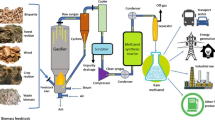
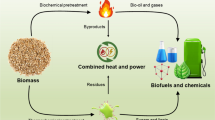
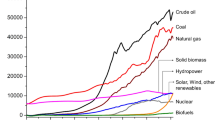
The 21st century’s goal to reduce CO2 emission is the driving force to replace petroleum with biomass. Although biomass has the potential to provide about ∼25% of global energy demands, biomass valorization (conversion to value-added products [VAPs]) also contributes to CO2 emissions. The use of microbial consortia rather than a single species is more efficient for biomass valorization. Thus, several molecular methods have been developed to decipher the composition of these consortia. Understanding the composition and diversity of microbes in a fermentation-based biomass valorization will enable the synthesis of artificial consortia to produce desirable products. Most works have identified the dominant microbial species in different biomass valorization processes and highlighted the influence of variables such as nanoparticles and fermentation inhibitors on microbial diversity and dynamics. The prominent microbial species in the microbial consortium are also discussed. This review will guide future works on microbial molecular profiling during biomass valorization.
Similar content being viewed by others
References
Jacquel, N., C.-W. Lo, Y.-H. Wei, H.-S. Wu, and S. S. Wang (2008) Isolation and purification of bacterial poly(3-hydroxyalkanoates). Biochem. Eng. J. 39: 15–27.
Moscovici, M. (2015) Present and future medical applications of microbial exopolysaccharides. Front. Microbiol. 6: 1012.
Rawoof, S. A. A., P. S. Kumar, D.-V. N. Vo, K. Devaraj, Y. Mani, T. Devaraj, and S. Subramanian (2021) Production of optically pure lactic acid by microbial fermentation: a review. Environ. Chem. Lett. 19: 539–556.
Vaishnav, N., A. Singh, M. Adsul, P. Dixit, S. K. Sandhu, A. Mathur, S. K. Puri, and R. R. Singhania (2018) Penicillium: the next emerging champion for cellulase production. Bioresour. Technol. Rep. 2: 131–140.
Verma, A., H. Singh, S. Anwar, A. Chattopadhyay, K. K. Tiwari, S. Kaur, and G. S. Dhilon (2017) Microbial keratinases: industrial enzymes with waste management potential. Crit. Rev. Biotechnol. 37: 476–491.
Mazzoli, R., F. Bosco, I. Mizrahi, E. A. Bayer, and E. Pessione (2014) Towards lactic acid bacteria-based biorefineries. Biotechnol. Adv. 32: 1216–1236.
Favaro, L., L. Alibardi, M. C. Lavagnolo, S. Casella, and M. Basaglia (2013) Effects of inoculum and indigenous microflora on hydrogen production from the organic fraction of municipal solid waste. Int. J. Hydrogen Energy. 38: 11774–11779.
Xu, L. and U. Tschirner (2011) Improved ethanol production from various carbohydrates through anaerobic thermophilic co-culture. Bioresour. Technol. 102: 10065–10071.
Xiros, C. and M. H. Studer (2017) A multispecies fungal biofilm approach to enhance the celluloyltic efficiency of membrane reactors for consolidated bioprocessing of plant biomass. Front. Microbiol. 8: 1930.
Minty, J. J., M. E. Singer, S. A. Scholz, C. H. Bae, J. H. Ahn, C. E. Foster, J. C. Liao, and X. N. Lin (2013) Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc. Natl. Acad. Sci. U. S. A. 110: 14592–14597.
Kalyani, D., K.-M. Lee, T.-S. Kim, J. Li, S. S. Dhiman, Y. C. Kang, and J.-K. Lee (2013) Microbial consortia for saccharification of woody biomass and ethanol fermentation. Fuel (Lond.) 107: 815–822.
Jawed, K., S. S. Yazdani, and M. A. Koffas (2019) Advances in the development and application of microbial consortia for metabolic engineering. Metab. Eng. Commun. 9: e00095. (Erratum published 2021, Metab. Eng. Commun. 13: e00186)
Albergaria, H. and N. Arneborg (2016) Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 100: 2035–2046.
Aquilanti, L., S. Santarelli, G. Silvestri, A. Osimani, A. Petruzzelli, and F. Clementi (2007) The microbial ecology of a typical Italian salami during its natural fermentation. Int. J. Food Microbiol. 120: 136–145.
ben Omar, N. and F. Ampe (2000) Microbial community dynamics during production of the Mexican fermented maize dough pozol. Appl. Environ. Microbiol. 66: 3664–3673.
Fontana, C., P. Sandro Cocconcelli, and G. Vignolo (2005) Monitoring the bacterial population dynamics during fermentation of artisanal Argentinean sausages. Int. J. Food Microbiol. 103: 131–142.
Mukisa, I. M., D. Porcellato, Y. B. Byaruhanga, C. M. B. K. Muyanja, K. Rudi, T. Langsrud, and J. A. Narvhus (2012) The dominant microbial community associated with fermentation of Obushera (sorghum and millet beverages) determined by culture-dependent and culture-independent methods. Int. J. Food Microbiol. 160: 1–10.
Cocolin, L., V. Alessandria, P. Dolci, R. Gorra, and K. Rantsiou (2013) Culture independent methods to assess the diversity and dynamics of microbiota during food fermentation. Int. J. Food Microbiol. 167: 29–43.
Botta, C. and L. Cocolin (2012) Microbial dynamics and biodiversity in table olive fermentation: culture-dependent and — independent approaches. Front. Microbiol. 3: 245.
Rossen, L., K. Holmstrøm, J. E. Olsen, and O. F. Rasmussen (1991) A rapid polymerase chain reaction (PCR)-based assay for the identification of Listeria monocytogenes in food samples. Int. J. Food Microbiol. 14: 145–151.
Randazzo, C. L., S. Torriani, A. D. L. Akkermans, W. M. de Vos, and E. E. Vaughan (2002) Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68: 1882–1892.
Alegría, A., P. Alvarez-Martín, N. Sacristán, E. Fernández, S. Delgado, and B. Mayo (2009) Diversity and evolution of the microbial populations during manufacture and ripening of Casín, a traditional Spanish, starter-free cheese made from cow’s milk. Int. J. Food Microbiol. 136: 44–51.
Wang, W., L. Yan, Z. Cui, Y. Gao, Y. Wang, and R. Jing (2011) Characterization of a microbial consortium capable of degrading lignocellulose. Bioresour. Technol. 102: 9321–9324.
Wang, X., H. Cui, J. Shi, X. Zhao, Y. Zhao, and Z. Wei (2015) Relationship between bacterial diversity and environmental parameters during composting of different raw materials. Bioresour. Technol. 198: 395–402.
Olsen, G. J., D. J. Lane, S. J. Giovannoni, N. R. Pace, and D. A. Stahl (1986) Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40: 337–365.
Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y.-H. Rogers, and H. O. Smith (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science. 304: 66–74.
Thomas, T., J. Gilbert, and F. Meyer (2012) Metagenomics — a guide from sampling to data analysis. Microb. Inform. Exp. 2: 3.
Ajayi-Banji, A. A., S. Rahman (2021) Efficacy of magnetite (Fe3O4) nanoparticles for enhancing solid-state anaerobic codigestion: Focus on reactor performance and retention time. Bioresour. Technol. 324: 124670.
Zerva, I., N. Remmas, and S. Ntougias (2019) Diversity and biotechnological potential of xylan-degrading microorganisms from orange juice processing waste. Water (Basel). 11: 274.
Portillo, M. and A. Mas (2016) Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding sequencing. Lebensm. Wiss. Technol. 72: 317–321.
Zapparoli, G., C. Reguant, A. Bordons, S. Torriani, and F. Dellaglio (2000) Genomic DNA fingerprinting of Oenococcus oeni strains by pulsed-field gel electrophoresis and randomly amplified polymorphic DNA-PCR. Curr. Microbiol. 40: 351–355.
Anyogu, A., B. Awamaria, J. P. Sutherland, and L. I. I. Ouoba (2014) Molecular characterisation and antimicrobial activity of bacteria associated with submerged lactic acid cassava fermentation. Food Control. 39: 119–127.
Peng, X., S. Zhang, L. Li, X. Zhao, Y. Ma, and D. Shi (2018) Long-term high-solids anaerobic digestion of food waste: Effects of ammonia on process performance and microbial community. Bioresour. Technol. 262: 148–158.
David, V., S. Terrat, K. Herzine, O. Claisse, S. Rousseaux, R. Tourdot-Maréchal, I. Masneuf-Pomarede, L. Ranjard, and H. Alexandre (2014) High-throughput sequencing of amplicons for monitoring yeast biodiversity in must and during alcoholic fermentation. J. Ind. Microbiol. Biotechnol. 41: 811–821.
González, Á., N. Hierro, M. Poblet, A. Mas, and J. M. Guillamón (2005) Application of molecular methods to demonstrate species and strain evolution of acetic acid bacteria population during wine production. Int. J. Food Microbiol. 102: 295–304.
de Melo Pereira, G. V., D. P. de Carvalho Neto, B. L. Maske, J. De Dea Lindner, A. S. Vale, G. R. Favero, J. Viesser, J. C. de Carvalho, A. Góes-Neto, and C. R. Soccol (2022) An updated review on bacterial community composition of traditional fermented milk products: what next-generation sequencing has revealed so far? Crit. Rev. Food Sci. Nutr. 62: 1870–1889.
Neefs, J.-M., Y. Van de Peer, P. De Rijk, S. Chapelle, and R. De Wachter (1993) Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 21: 3025–3049.
Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field (1990) Genetic diversity in Sargasso Sea bacterioplankton. Nature. 345: 60–63.
Ros, M., J. de Souza Oliveira Filho, M. D. Perez Murcia, M. A. Bustamante, R. Moral, M. D. Coll, A. B. Lopez Santisima-Trinidad, and J. A. Pascual (2017) Mesophilic anaerobic digestion of pig slurry and fruit and vegetable waste: dissection of the microbial community structure. J. Clean. Prod. 156: 757–765.
Sahoo, R. K., E. Subudhi, and M. Kumar (2014) Quantitative approach to track lipase producing Pseudomonas sp. S1 in nonsterilized solid state fermentation. Lett. Appl. Microbiol. 58: 610–616.
Caporaso, J. G., J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Peña, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelley, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevinsky, P. J. Turnbaugh, W. A. Walters, J. Widmann, T. Yatsunenko, J. Zaneveld, and R. Knight (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7: 335–336.
Edgar, R. C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26: 2460–2461.
Schloss, P. D., S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn, and C. F. Weber (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75: 7537–7541.
Claesson, M. J., Q. Wang, O. O’Sullivan, R. Greene-Diniz, J. R. Cole, R. P. Ross, and P. W. O’Toole (2010) Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38: e200.
Hugon, P., J.-C. Lagier, C. Robert, C. Lepolard, L. Papazian, D. Musso, B. Vialettes, and D. Raoult (2013) Molecular studies neglect apparently gram-negative populations in the human gut microbiota. J. Clin. Microbiol. 51: 3286–3293.
Liu, Z., C. Lozupone, M. Hamady, F. D. Bushman, and R. Knight (2007) Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35: e120.
Kostinek, M., I. Specht, V. A. Edward, C. Pinto, M. Egounlety, C. Sossa, S. Mbugua, C. Dortu, P. Thonart, L. Taljaard, M. Mengu, C. M. Franz, and W. H. Holzapfel (2007) Characterisation and biochemical properties of predominant lactic acid bacteria from fermenting cassava for selection as starter cultures. Int. J. Food Microbiol. 114: 342–351.
Plengvidhya, V., F. BreidtJr., Z. Lu, and H. P. Fleming (2007) DNA fingerprinting of lactic acid bacteria in sauerkraut fermentations. Appl. Environ. Microbiol. 73: 7697–7702.
Vouidibio Mbozo, A. B., S. C. Kobawila, A. Anyogu, B. Awamaria, D. Louembe, J. P. Sutherland, and L. I. Ouoba (2017) Investigation of the diversity and safety of the predominant Bacillus pumilus sensu lato and other Bacillus species involved in the alkaline fermentation of cassava leaves for the production of Ntoba Mbodi. Food Control. 82: 154–162.
Ampe, F., N. ben Omar, C. Moizan, C. Wacher, and J. P. Guyot (1999) Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65: 5464–5473.
Hamad, S. H., M. C. Dieng, M. A. Ehrmann, and R. F. Vogel (1997) Characterization of the bacterial flora of Sudanese sorghum flour and sorghum sourdough. J. Appl. Microbiol. 83: 764–770.
Quast, C., E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies, and F. O. Glöckner (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41: D590–D596.
Compeau, P. E. C., P. A. Pevzner, and G. Tesler (2011) How to apply de Bruijn graphs to genome assembly. Nat. Biotechnol. 29: 987–991.
Chen, Y., J. Sheng, T. Jiang, J. Stevens, X. Feng, and N. Wei (2016) Transcriptional profiling reveals molecular basis and novel genetic targets for improved resistance to multiple fermentation inhibitors in Saccharomyces cerevisiae. Biotechnol. Biofuels. 9: 9.
Verce, M., J. Schoonejans, C. Hernandez Aguirre, R. Molina-Bravo, L. De Vuyst, and S. Weckx (2021) A combined metagenomics and metatranscriptomics approach to unravel Costa Rican cocoa box fermentation processes reveals yet unreported microbial species and functionalities. Front. Microbiol. 12: 641185.
Ercolini, D. (2013) High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 79: 3148–3155.
Erkus, O., V. C. L. de Jager, R. T. C. M. Geene, I. van Alen-Boerrigter, L. Hazelwood, S. A. F. T. van Hijum, M. Kleerebezem, and E. J. Smid (2016) Use of propidium monoazide for selective profiling of viable microbial cells during Gouda cheese ripening. Int. J. Food Microbiol. 228: 1–9.
Simmons, C. W., A. P. Reddy, P. D’haeseleer, J. Khudyakov, K. Billis, A. Pati, B. A. Simmons, S. W. Singer, M. P. Thelen, and J. S. VanderGheynst (2014) Metatranscriptomic analysis of lignocellulolytic microbial communities involved in high-solids decomposition of rice straw. Biotechnol. Biofuels. 7: 495.
Alessi, A. M., S. M. Bird, N. C. Oates, Y. Li, A. A. Dowle, E. H. Novotny, E. R. deAzevedo, J. P. Bennett, I. Polikarpov, J. Young, S. J. McQueen-Mason, and N. C. Bruce (2018) Defining functional diversity for lignocellulose degradation in a microbial community using multi-omics studies. Biotechnol. Biofuels. 11: 166.
He, B., S. Jin, J. Cao, L. Mi, and J. Wang (2019) Metatranscriptomics of the Hu sheep rumen microbiome reveals novel cellulases. Biotechnol. Biofuels. 12: 153.
Muyzer, G., E. C. de Waal, and A. G. Uitterlinden (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59: 695–700.
Cocolin, L., M. Manzano, C. Cantoni, and G. Comi (2001) Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67: 5113–5121.
Masoud, W., L. B. Cesar, L. Jespersen, and M. Jakobsen (2004) Yeast involved in fermentation of Coffea arabica in East Africa determined by genotyping and by direct denaturating gradient gel electrophoresis. Yeast. 21: 549–556.
Wei, Q., H. Wang, Z. Chen, Z. Lv, Y. Xie, and F. Lu (2013) Profiling of dynamic changes in the microbial community during the soy sauce fermentation process. Appl. Microbiol. Biotechnol. 97: 9111–9119.
Cocolin, L., L. F. Bisson, and D. A. Mills (2000) Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 189: 81–87.
El Sheikha, A. F. (2019) Molecular detection of mycotoxigenic fungi in foods: the case for using PCR-DGGE. Food Biotechnol. 33: 54–108.
Zhang, W., Y. Mo, J. Yang, J. Zhou, Y. Lin, A. Isabwe, J. Zhang, X. Gao, and Z. Yu (2018) Genetic diversity pattern of microeukaryotic communities and its relationship with the environment based on PCR-DGGE and T-RFLP techniques in Dongshan Bay, southeast China. Cont. Shelf Res. 164: 1–9.
Han, R., Y. Yuan, Q. Cao, Q. Li, L. Chen, D. Zhu, and D. Liu (2018) PCR-DGGE analysis on microbial community structure of rural household biogas digesters in Qinghai Plateau. Curr. Microbiol. 75: 541–549.
Salvachúa, D., A. Z. Werner, I. Pardo, M. Michalska, B. A. Black, B. S. Donohoe, S. J. Haugen, R. Katahira, S. Notonier, K. J. Ramirez, A. Amore, S. O. Purvine, E. M. Zink, P. E. Abraham, R. J. Giannone, S. Poudel, P. D. Laible, R. L. Hettich, and G. T. Beckham (2020) Outer membrane vesicles catabolize lignin-derived aromatic compounds in Pseudomonas putida KT2440. Proc. Natl. Acad. Sci. U. S. A. 117: 9302–9310.
Cleary, D. F. R., K. Smalla, L. C. S. Mendonça-Hagler, and N. C. M. Gomes (2012) Assessment of variation in bacterial composition among microhabitats in a mangrove environment using DGGE fingerprints and barcoded pyrosequencing. PLoS One. 7: e29380.
Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams (1996) Real time quantitative PCR. Genome Res. 6: 986–994.
De Vuyst, L., N. Camu, T. De Winter, K. Vandemeulebroecke, V. Van de Perre, M. Vancanneyt, P. De Vos, and I. Cleenwerck (2008) Validation of the (GTG)(5)-rep-PCR fingerprinting technique for rapid classification and identification of acetic acid bacteria, with a focus on isolates from Ghanaian fermented cocoa beans. Int. J. Food Microbiol. 125: 79–90.
Kumar, M., A. Joshi, R. Kashyap, and S. Khanna (2011) Production of xylanase by Promicromonospora sp MARS with rice straw under non sterile conditions. Process Biochem. 46: 1614–1618.
Paludan-Müller, C., R. Valyasevi, H. H. Huss, and L. Gram (2002) Genotypic and phenotypic characterization of garlic-fermenting lactic acid bacteria isolated from som-fak, a Thai low-salt fermented fish product. J. Appl. Microbiol. 92: 307–314.
Wu, Y. R. and J. He (2013) Characterization of anaerobic consortia coupled lignin depolymerization with biomethane generation. Bioresour. Technol. 139: 5–12.
de Oliveira, C. T., L. Pellenz, J. Q. Pereira, A. Brandelli, and D. J. Daroit (2016) Screening of bacteria for protease production and feather degradation. Waste Biomass Valorization. 7: 447–453.
Joblin, K. N., G. E. Naylor, and A. G. Williams (1990) Effect of Methanobrevibacter smithii on xylanolytic activity of anaerobic ruminal fungi. Appl. Environ. Microbiol. 56: 2287–2295.
Bader, J., E. Mast-Gerlach, M. K. Popović, R. Bajpai, and U. Stahl (2010) Relevance of microbial coculture fermentations in biotechnology. J. Appl. Microbiol. 109: 371–387.
Pandhal, J. and J. Noirel (2014) Synthetic microbial ecosystems for biotechnology. Biotechnol. Lett. 36: 1141–1151.
Joblin, K. N., H. Matsui, G. E. Naylor, and K. Ushida (2002) Degradation of fresh ryegrass by methanogenic co-cultures of ruminal fungi grown in the presence or absence of Fibrobacter succinogenes. Curr. Microbiol. 45: 46–53.
Cheng, X.-Y. and C.-Z. Liu (2012) Fungal pretreatment enhances hydrogen production via thermophilic fermentation of cornstalk. Appl. Energy. 91: 1–6.
Pessiot, J., R. Nouaille, M. Jobard, R. R. Singhania, A. Bournilhas, G. Christophe, P. Fontanille, P. Peyret, G. Fonty, and C. Larroche (2012) Fed-batch anaerobic valorization of slaughterhouse by-products with mesophilic microbial consortia without methane production. Appl. Biochem. Biotechnol. 167: 1728–1743.
Zhang, J., J. Liu, Z. Shi, L. Liu, and J. Chen (2010) Manipulation of B. megaterium growth for efficient 2-KLG production by K. vulgare. Process Biochem. 45: 602–606.
Liu, Y., M. Ding, W. Ling, Y. Yang, X. Zhou, B.-Z. Li, T. Chen, Y. Nie, M. Wang, B. Zeng, X. Li, H. Liu, B. Sun, H. Xu, J. Zhang, Y. Jiao, Y. Hou, H. Yang, S. Xiao, Q. Lin, X. He, W. Liao, Z. Jin, Y. Xie, B. Zhang, T. Li, X. Lu, J. Li, F. Zhang, X.-L. Wu, H. Song, and Y.-J. Yuan (2017) A three-species microbial consortium for power generation. Energy Environ. Sci. 10: 1600–1609.
Shahab, R. L., S. Brethauer, M. P. Davey, A. G. Smith, S. Vignolini, J. S. Luterbacher, and M. H. Studer (2020) A heterogeneous microbial consortium producing short-chain fatty acids from lignocellulose. Science. 369: eabb1214.
Poszytek, K., M. Ciezkowska, A. Sklodowska, and L. Drewniak (2016) Microbial Consortium with High Cellulolytic Activity (MCHCA) for enhanced biogas production. Front. Microbiol. 7: 324.
Sun, Y., Z. Xu, Y. Zheng, J. Zhou, and Z. Xiu (2019) Efficient production of lactic acid from sugarcane molasses by a newly microbial consortium CEE-DL15. Process Biochem. 81: 132–138.
Schwalm, N. D., III, W. Mojadedi, E. S. Gerlach, M. Benyamin, M. A. Perisin, and K. L. Akingbade (2019) Developing a microbial consortium for enhanced metabolite production from simulated food waste. Fermentation (Basel) 5: 98.
Zhou, K., K. Qiao, S. Edgar, and G. Stephanopoulos (2015) Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 33: 377–383.
Zhao, C., J. P. Sinumvayo, Y. Zhang, and Y. Li (2019) Design and development of a “Y-shaped” microbial consortium capable of simultaneously utilizing biomass sugars for efficient production of butanol. Metab. Eng. 55: 111–119.
Chen, Y., Z. Yang, Y. Zhang, Y. Xiang, R. Xu, M. Jia, J. Cao, and W. Xiong (2020) Effects of different conductive nanomaterials on anaerobic digestion process and microbial community of sludge. Bioresour. Technol. 304: 123016.
Ajay, C. M., S. Mohan, P. Dinesha, and M. A. Rosen (2020) Review of impact of nanoparticle additives on anaerobic digestion and methane generation. Fuel (Lond.) 277: 118234.
Huangfu, X., Y. Xu, C. Liu, Q. He, J. Ma, C. Ma, and R. Huang (2019) A review on the interactions between engineered nanoparticles with extracellular and intracellular polymeric substances from wastewater treatment aggregates. Chemosphere. 219: 766–783.
Zhang, J., Z. Wang, T. Lu, J. Liu, Y. Wang, P. Shen, and Y. Wei (2019) Response and mechanisms of the performance and fate of antibiotic resistance genes to nano-magnetite during anaerobic digestion of swine manure. J. Hazard. Mater. 366: 192–201.
Jing, Y., J. Wan, I. Angelidaki, S. Zhang, and G. Luo (2017) iTRAQ quantitative proteomic analysis reveals the pathways for methanation of propionate facilitated by magnetite. Water Res. 108: 212–221.
Baek, G., J. Kim, K. Cho, H. Bae, and C. Lee (2015) The biostimulation of anaerobic digestion with (semi)conductive ferric oxides: their potential for enhanced biomethanation. Appl. Microbiol. Biotechnol. 99: 10355–10366.
Yin, Q., J. Miao, B. Li, and G. Wu (2017) Enhancing electron transfer by ferroferric oxide during the anaerobic treatment of synthetic wastewater with mixed organic carbon. Int. Biodeterior. Biodegradation. 119: 104–110.
Abdelsalam, E., M. Samer, Y. A. Attia, M. A. Abdel-Hadi, H. E. Hassan, and Y. Badr (2017) Influence of zero valent iron nanoparticles and magnetic iron oxide nanoparticles on biogas and methane production from anaerobic digestion of manure. Energy (Oxf.) 120: 842–853.
Kökdemir Ünşar, E. and N. A. Perendeci (2018) What kind of effects do Fe2O3 and Al2O3 nanoparticles have on anaerobic digestion, inhibition or enhancement? Chemosphere. 211: 726–735.
Zhao, Z., Y. Zhang, Y. Li, X. Quan, and Z. Zhao (2018) Comparing the mechanisms of ZVI and Fe3O4 for promoting waste-activated sludge digestion. Water Res. 144: 126–133.
Baral, N. R. and A. Shah (2014) Microbial inhibitors: formation and effects on acetone-butanol-ethanol fermentation of lignocellulosic biomass. Appl. Microbiol. Biotechnol. 98: 9151–9172.
Tian, H., I. A. Fotidis, E. Mancini, L. Treu, A. Mahdy, M. Ballesteros, C. González-Fernández, and I. Angelidaki (2018) Acclimation to extremely high ammonia levels in continuous biomethanation process and the associated microbial community dynamics. Bioresour. Technol. 247: 616–623.
Wang, F. Q., H. Xie, W. Chen, E. T. Wang, F. G. Du, and A. D. Song (2013) Biological pretreatment of corn stover with ligninolytic enzyme for high efficient enzymatic hydrolysis. Bioresour. Technol. 144: 572–578.
Sasano, Y., D. Watanabe, K. Ukibe, T. Inai, I. Ohtsu, H. Shimoi, and H. Takagi (2012) Overexpression of the yeast transcription activator Msn2 confers furfural resistance and increases the initial fermentation rate in ethanol production. J. Biosci. Bioeng. 113: 451–455.
Kim, D. and J.-S. Hahn (2013) Roles of the Yap1 transcription factor and antioxidants in Saccharomyces cerevisiae’s tolerance to furfural and 5-hydroxymethylfurfural, which function as thiol-reactive electrophiles generating oxidative stress. Appl. Environ. Microbiol. 79: 5069–5077.
Conway, J. M., B. S. McKinley, N. L. Seals, D. Hernandez, P. A. Khatibi, S. Poudel, R. J. Giannone, R. L. Hettich, A. M. Williams-Rhaesa, G. L. Lipscomb, M. Adams, and R. M. Kelly (2017) Functional analysis of the glucan degradation locus in Caldicellulosiruptor bescii reveals essential roles of component glycoside hydrolases in plant biomass deconstruction. Appl. Environ. Microbiol. 83: e01828–17.
Peng, X., W. Qiao, S. Mi, X. Jia, H. Su, and Y. Han (2015) Characterization of hemicellulase and cellulase from the extremely thermophilic bacterium Caldicellulosiruptor owensensis and their potential application for bioconversion of lignocellulosic biomass without pretreatment. Biotechnol. Biofuels. 8: 131.
Lochner, A., R. J. Giannone, M. RodriguezJr., M. B. Shah, J. R. Mielenz, M. Keller, G. Antranikian, D. E. Graham, and R. L. Hettich (2011) Use of label-free quantitative proteomics to distinguish the secreted cellulolytic systems of Caldicellulosiruptor bescii and Caldicellulosiruptor obsidiansis. Appl. Environ. Microbiol. 77: 4042–4054.
Brunecky, R., M. Alahuhta, Q. Xu, B. S. Donohoe, M. F. Crowley, I. A. Kataeva, S. J. Yang, M. G. Resch, M. W. Adams, V. V. Lunin, M. E. Himmel, and Y. J. Bomble (2013) Revealing nature’s cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science. 342: 1513–1516.
Bing, R. G., C. T. Straub, D. B. Sulis, J. P. Wang, M. W. W. Adams, and R. M. Kelly (2022) Plant biomass fermentation by the extreme thermophile Caldicellulosiruptor bescii for coproduction of green hydrogen and acetone: technoeconomic analysis. Bioresour. Technol. 348: 126780.
Ali, N., H. I. Hamouda, H. Su, J. Feng, Z.-Y. Liu, M. Lu, and F.-L. Li (2020) A two-stage anaerobic bioconversion of corn stover: impact of pure bacterial pretreatment on methane production. Environ. Technol. Innov. 20: 101141.
Hamilton-Brehm, S. D., J. J. Mosher, T. Vishnivetskaya, M. Podar, S. Carroll, S. Allman, T. J. Phelps, M. Keller, and J. G. Elkins (2010) Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl. Environ. Microbiol. 76: 1014–1020.
Kataeva, I., M. B. Foston, S.-J. Yang, S. Pattathil, A. K. Biswal, F. L. PooleII, M. Basen, A. M. Rhaesa, T. P. Thomas, P. Azadi, V. Olman, T. D. Saffold, K. E. Mohler, D. L. Lewis, C. Doeppke, Y. Zeng, T. J. Tschaplinski, W. S. York, M. Davis, D. Mohnen, Y. Xu, A. J. Ragauskas, S.-Y. Ding, R. M. Kelly, M. G. Hahn, and M. W. W. Adams (2013) Carbohydrate and lignin are simultaneously solubilized from unpretreated switchgrass by microbial action at high temperature. Energy Environ. Sci. 6: 2186–2195.
del Cerro, C., E. Erickson, T. Dong, A. R. Wong, E. K. Eder, S. O. Purvine, H. D. Mitchell, K. K. Weitz, L. M. Markillie, M. C. Burnet, D. W. Hoyt, R. K. Chu, J. F. Cheng, K. J. Ramirez, R. Katahira, W. Xiong, M. E. Himmel, V. Subramanian, J. G. Linger, and D. Salvachúa (2021) Intracellular pathways for lignin catabolism in white-rot fungi. Proc. Natl. Acad. Sci. U. S. A. 118: e2017381118.
Coelho-Moreira, J. D. S., G. M. Maciel, R. Castoldi, S. D. S. Mariano, F. D. Inácio, A. Bracht, and R. M. Peralta (2013) Involvement of lignin-modifying enzymes in the degradation of herbicides. pp. 165–187. In: A. J. Price and J. A. Kelton (eds.). Herbicides — Advances in Research. IntechOpen, London, UK.
Vasco-Correa, J. and Y. Li (2015) Solid-state anaerobic digestion of fungal pretreated Miscanthus sinensis harvested in two different seasons. Bioresour. Technol. 185: 211–217.
Zhao, J., X. Ge, J. Vasco-Correa, and Y. Li (2014) Fungal pretreatment of unsterilized yard trimmings for enhanced methane production by solid-state anaerobic digestion. Bioresour. Technol. 158: 248–252.
Akyol, Ç., O. Ince, M. Bozan, E. G. Ozbayram, and B. Ince (2019) Biological pretreatment with Trametes versicolor to enhance methane production from lignocellulosic biomass: a metagenomic approach. Ind. Crops Prod. 140: 111659.
Mustafa, A. M., T. G. Poulsen, and K. Sheng (2016) Fungal pretreatment of rice straw with Pleurotus ostreatus and Trichoderma reesei to enhance methane production under solidstate anaerobic digestion. Appl. Energy. 180: 661–671.
Yadav, M., K. Paritosh, N. Pareek, and V. Vivekanand (2019) Coupled treatment of lignocellulosic agricultural residues for augmented biomethanation. J. Clean. Prod. 213: 75–88.
González, C., Y. Wu, A. Zuleta-Correa, G. Jaramillo, and J. Vasco-Correa (2021) Biomass to value-added products using microbial consortia with white-rot fungi. Bioresour. Technol. Rep. 16: 100831.
Wang, W., T. Yuan, and B. Cui (2014) Biological pretreatment with white rot fungi and their co-culture to overcome lignocellulosic recalcitrance for improved enzymatic digestion. Bioresources. 9: 3968–3976.
Wang, R., T. You, G. Yang, and F. Xu (2017) Efficient short time white rot—brown rot fungal pretreatments for the enhancement of enzymatic saccharification of corn cobs. ACS Sustain. Chem. Eng. 5: 10849–10857.
Ma, K. and Z. Ruan (2015) Production of a lignocellulolytic enzyme system for simultaneous bio-delignification and saccharification of corn stover employing co-culture of fungi. Bioresour. Technol. 175: 586–593.
Hermosilla, E., O. Rubilar, H. Schalchli, A. S. da Silva, V. Ferreira-Leitao, and M. C. Diez (2018) Sequential white-rot and brown-rot fungal pretreatment of wheat straw as a promising alternative for complementary mild treatments. Waste Manag. 79: 240–250.
Xie, P., L. Fan, L. Huang, and C. Zhang (2020) An innovative co-fungal treatment to poplar bark sawdust for delignification and polyphenol enrichment. Ind. Crops Prod. 157: 112896.
Yoon, L. W., G. C. Ngoh, A. S. M. Chua, M. F. Abdul Patah, and W. H. Teoh (2019) Process intensification of cellulase and bioethanol production from sugarcane bagasse via an integrated saccharification and fermentation process. Chem. Eng. Process. 142: 107528.
Horisawa, S., A. Inoue, and Y. Yamanaka (2019) Direct ethanol production from lignocellulosic materials by mixed culture of wood rot fungi Schizophyllum commune, Bjerkandera adusta, and Fomitopsis palustris. Fermentation (Basel). 5: 21.
Horisawa, S., H. Ando, O. Ariga, and Y. Sakuma (2015) Direct ethanol production from cellulosic materials by consolidated biological processing using the wood rot fungus Schizophyllum commune. Bioresour. Technol. 197: 37–41.
Tri, C. L. and I. Kamei (2020) Butanol production from cellulosic material by anaerobic co-culture of white-rot fungus Phlebia and bacterium Clostridium in consolidated bioprocessing. Bioresour. Technol. 305: 123065.
Biderre-Petit, C., D. Jézéquel, E. Dugat-Bony, F. Lopes, J. Kuever, G. Borrel, E. Viollier, G. Fonty, and P. Peyret (2011) Identification of microbial communities involved in the methane cycle of a freshwater meromictic lake. FEMS Microbiol. Ecol. 77: 533–545.
Kumar, M., S. You, J. Beiyuan, G. Luo, J. Gupta, S. Kumar, L. Singh, S. Zhang, and D. C. W. Tsang (2021) Lignin valorization by bacterial genus Pseudomonas: state-of-the-art review and prospects. Bioresour. Technol. 320: 124412.
Elmore, J. R., G. N. Dexter, D. Salvachúa, M. O’Brien, D. M. Klingeman, K. Gorday, J. K. Michener, D. J. Peterson, G. T. Beckham, and A. M. Guss (2020) Engineered Pseudomonas putida simultaneously catabolizes five major components of corn stover lignocellulose: glucose, xylose, arabinose, p-coumaric acid, and acetic acid. Metab. Eng. 62: 62–71.
Lee, S., J.-H. Sohn, J.-H. Bae, S. C. Kim, and B. H. Sung (2020) Current status of Pseudomonas putida engineering for lignin valorization. Biotechnol. Bioprocess Eng. 25: 862–871.
Yang, C., F. Yue, Y. Cui, Y. Xu, Y. Shan, B. Liu, Y. Zhou, and X. Lü (2018) Biodegradation of lignin by Pseudomonas sp. Q18 and the characterization of a novel bacterial DyP-type peroxidase. J. Ind. Microbiol. Biotechnol. 45: 913–927.
Ghosh, T., T.-D. Ngo, A. Kumar, C. Ayranci, and T. Tang (2019) Cleaning carbohydrate impurities from lignin using Pseudomonas fluorescens. Green Chem. 21: 1648–1659.
Wang, X., L. Lin, J. Dong, W. Wang, H. Wang, Z. Zhang, and X. Yu (2018) Simultaneous improvements of Pseudomonas cell growth and polyhydroxyalkanoate production from a lignin derivative for lignin-consolidated bioprocessing. Appl. Environ. Microbiol. 84: e01469–18.
Nikel, P. I. and V. de Lorenzo (2018) Pseudomonas putida as a functional chassis for industrial biocatalysis: from native biochemistry to trans-metabolism. Metab. Eng. 50: 142–155.
Zerva, I., N. Remmas, P. Melidis, G. Sylaios, P. Stathopoulou, G. Tsiamis, and S. Ntougias (2022) Biotreatment, microbial community structure and valorization potential of pepper processing wastewater in an immobilized cell bioreactor. Waste and Biomass Valor. 13: 1431–1447.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Adeniyi, A., Bello, I., Mukaila, T. et al. A Review of Microbial Molecular Profiling during Biomass Valorization. Biotechnol Bioproc E 27, 515–532 (2022). https://doi.org/10.1007/s12257-022-0026-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-022-0026-8