Abstract
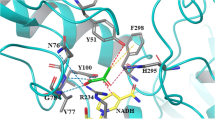
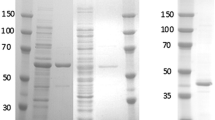
Adipic acid is an important monomer for the production of nylon-6,6 polyamide. One novel biological route for the synthesis of adipic acid, which combines the lysine synthetic pathway and glutaconic acid production pathway, has been suggested, but this route has suffered from the lack of an efficient 2-oxoadipate reductase connecting the two pathways or converting 2-oxoadipate to 2-hydroxyadipate. In this study, we report that the lactate dehydrogenase of Alcaligenes eutrophus H16 is a promising catalyst for this reaction. The lactate dehydrogenase gene (Ae-ldhO) was cloned, expressed in Escherichia coli, purified, and characterized. The recombinant enzyme, having a molecular weight of 36.7 kDa, exhibited broad substrate specificity for various 2-oxoacids. NADH was the preferred coenzyme over NADPH for all 2-oxoacids tested. The maximum specific activity of Ae-LdhO on 2-oxoadipate was 454.5 ± 20.1 U/mg protein at pH 7.0 and 30℃. The K m values for 2-oxoadipic acid and NADH were 0.32 ± 0.02 and 0.09 ± 0.002 mM, respectively. The activity of Ae-LdhO was enhanced in the presence of some metal ions, such as Mg2+, Co2+ or Ni2+, whereas it was completely inhibited by Hg2+, Ag+, Cu2+ and DTT.
Similar content being viewed by others
References
Polen, T., M. Spelberg, and M. Bott (2013) Toward biotechnological production of adipic acid and precursors from biorenewables. J. Biotechnol. 167: 75–84.
Merchant, R. and L. Consulting (2011) Adipic acid 2011 World Market Outlook and Forecast.
Musser, M. T. (2005) Adipic acid. ULLMANN’S Encyclopedia of Industrial Chemistry. pp. 537–548. vol. 1. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Ince, W. H. (1895) Preparation of adipic acid and some of its derivatives. J. Chem. Soc. Trans. 67: 155–159.
Usui, Y. and K. Sato (2003) A green method of adipic acid synthesis: Organic solvent- and halide-free oxidation of cycloalkanones with 30% hydrogen peroxide. Green Chem. 5: 373–375.
Cao, B., A. Geng, and K. C. Loh (2008) Induction of ortho- and meta-cleavage pathways in Pseudomonas in biodegradation of high benzoate concentration: MS identification of catabolic enzymes. Appl. Microbiol. Biotechnol. 81: 99–107.
Collier, L. S., G. L. Gaines, and E. L. Neidle (1998) Regulation of benzoate degradation in Acinetobacter sp. strain ADP1 by BenM, a LysR-type transcriptional activator. J. Bacteriol. 180: 2493–2501.
Denef, V. J., J. A. Klappenbach, M. A. Patrauchan, C. Florizone, J. L. Rodrigues, T. V. Tsoi, W. Verstraete, L. D. Eltis, and J. M. Tiedje (2006) Genetic and genomic insights into the role of benzoate-catabolic pathway redundancy in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 72: 585–595.
Jeffrey, W. H., S. M. Cuskey, P. J. Chapman, S. Resnick, and R. H. Olsen (1992) Characterization of Pseudomonas putida mutants unable to catabolize benzoate: Cloning and characterization of Pseudomonas genes involved in benzoate catabolism and isolation of a chromosomal DNA fragment able to substitute for xylS in activation of the TOL lower-pathway promoter. J. Bacteriol. 174: 4986–4996.
Kitagawa, W., K. Miyauchi, E. Masai, and M. Fukuda (2001) Cloning and characterization of benzoate catabolic genes in the gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. J. Bacteriol. 183: 6598–6606.
McFall, S. M., S. A. Chugani, and A. M. Chakrabarty (1998) Transcriptional activation of the catechol and chlorocatechol operons: Variations on a theme. Gene. 223: 257–267.
Moreno, R. and F. Rojo (2008) The Target for the Pseudomonas putida Crc global regulator in the benzoate degradation pathway is the benr transcriptional regulator. J. Bacteriol. 190: 1539–1545.
Neidle, E. L., M. K. Shapiro, and L. N. Ornston (1987) Cloning and expression in Escherichia coli of Acinetobacter calcoaceticus genes for benzoate degradation. J. Bacteriol. 169: 5496–5503.
Takenaka, S., E. Setyorini, Y. J. Kim, S. Murakami, and K. Aoki (2005) Constitutive synthesis of enzymes involved in 2-aminophenol metabolism and inducible synthesis of enzymes involved in benzoate, p-hydroxybenzoate, and protocatechuate metabolism in Pseudomonas sp. strain AP-3. Biosci. Biotechnol. Biochem. 69: 1033–1035.
Yoon, Y. H., S. H. Yun, S. H. Park, S. Y. Seol, S. H. Leem, and S. I. Kim (2007) Characterization of a new catechol branch of the beta-ketoadipate pathway induced for benzoate degradation in Acinetobacter lwoffii K24. Biochem. Biophys. Res. Commun. 360: 513–519.
Zhan, Y., H. Yu, Y. Yan, M. Chen, W. Lu, S. Li, Z. Peng, W. Zhang, S. Ping, J. Wang, and M. Lin (2008) Genes involved in the benzoate catabolic pathway in Acinetobacter calcoaceticus PHEA-2. Curr. Microbiol. 57: 609–614.
De, G. D. (2010) Bio-adipic acid prepares for entry. pp: 22–23. ICIS Chemical Business. September 27–October 3.
Niu, W., K. M. Draths, and J. W. Frost (2002) Benzene-free synthesis of adipic acid. Biotechnol. Prog. 18: 201–211.
Thomas, J. M., R. Raja, B. F. Johnson, T. J. O'Connell, G. Sankar, and T. Khimyak (2003) Bimetallic nanocatalysts for the conversion of muconic acid to adipic acid. Chem. Commun. (Camb). 1126–1127.
Parthasarathy, A., A. J. Pierik, J. Kahnt, O. Zelder, and W. Buckel (2011) Substrate specificity of 2-hydroxyglutaryl-CoA dehydratase from Clostridium symbiosum: Toward a bio-based production of adipic acid. Biochem. 50: 3540–3550.
Steinbuchel, A. and H. G. Schlegel (1983) NAD-linked L(+)-lactate dehydrogenase from the strict aerobe Alcaligenes eutrophus. 1. Purification and properties. Eur. J. Biochem. 130: 321–328.
Sambrook, J. and D. Russell (2001) Molecular cloning: A Laboratory Manual. 3rd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA.
Laemmli, U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685.
Bradford, M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254.
Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman (1990) Basic local alignment search tool. J. Mol. Biol. 215: 403–410.
Eswar, N., B. Webb, M. A. Marti-Renom, M. S. Madhusudhan, D. Eramian, M. Y. Shen, U. Pieper, and A. Sali (2007) Comparative protein structure modeling using MODELLER. Curr. Protoc Protein Sci. 15: 5.6.1–5.6.30.
Shen, M. Y. and A. Sali (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci. 15: 2507–2524.
Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton (1993) PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283–291.
Eisenberg, D., R. Luthy, and J. U. Bowie (1997) VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 277: 396–404.
Van Der Spoel, D., E. Lindahl, B. Hess, G. Groenhof, A. E. Mark, and H. J. Berendsen (2005) GROMACS: Fast, flexible, and free. J. Comput. Chem. 26: 1701–1718.
Lee, P., S. Raj, S. Zhou, S. Ashok, S. Edwardraja, and S. Park (2014) 3-hydroxyisobutyrate dehydrogenase-I from Pseudomonas denitrificans ATCC 13867 degrades 3-hydroxypropionic acid. Biotechnol. Bioproc. Eng. 19: 1–7.
Sokalingam, S., B. Madan, G. Raghunathan, and S.-G. Lee (2013) In silico study on the effect of surface lysines and arginines on the electrostatic interactions and protein stability. Biotechnol. Bioproc. Eng. 18: 18–26.
Morris, G. M., R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell, and A. J. Olson (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30: 2785–2791.
Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne (2000) The protein data bank. Nucleic Acids Res. 28: 235–242.
Porter, C. T., G. J. Bartlett, and J. M. Thornton (2004) The catalytic site Atlas: A resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 32: 129–133.
Raj, S., C. Rathnasingh, W.-C. Jung, E. Selvakumar, and S. Park (2010) A Novel NAD+-dependent aldehyde dehydrogenase encoded by the puuC gene of Klebsiella pneumoniae DSM 2026 that utilizes 3-hydroxypropionaldehyde as a substrate. Biotechnol. Bioproc. Eng. 15: 131–138.
Jo, J. E., S. Mohan Raj, C. Rathnasingh, E. Selvakumar, W. C. Jung, and S. Park (2008) Cloning, expression, and characterization of an aldehyde dehydrogenase from Escherichia coli K-12 that utilizes 3-Hydroxypropionaldehyde as a substrate. Appl. Microbiol. Biotechnol. 81: 51–60.
Berrios-Rivera, S. J., G. N. Bennett, and K. Y. San (2002) Metabolic engineering of Escherichia coli: Increase of NADH availability by overexpressing an NAD(+)-dependent formate dehydrogenase. Metab. Eng. 4: 217–229.
Schwartz, E., U. Gerischer, and B. Friedrich (1998) Transcriptional regulation of Alcaligenes eutrophus hydrogenase genes. J. Bacteriol. 180: 3197–3204.
Garvie, E. I. (1980) Bacterial lactate dehydrogenases. Microbiol. Rev. 44: 106–139.
Kabsch, W. and C. Sander (1983) Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopoly. 22: 2577–2637.
Rossmann, M. G. and P. Argos (1976) Exploring structural homology of proteins. J. Mol. Biol. 105: 75–95.
Eriksson, M., U. Uhlin, S. Ramaswamy, M. Ekberg, K. Regnstrom, B. M. Sjoberg, and H. Eklund (1997) Binding of allosteric effectors to ribonucleotide reductase protein R1: Reduction of active-site cysteines promotes substrate binding. Structure. 5: 1077–1092.
Forouhar, F., I. Lee, J. Benach, K. Kulkarni, R. Xiao, T. B. Acton, G. T. Montelione, and L. Tong (2004) A novel NAD-binding protein revealed by the crystal structure of 2,3-diketo-L-gulonate reductase (YiaK). J. Biol. Chem. 279: 13148–13155.
Author information
Authors and Affiliations
Corresponding author
Additional information
These two author’s contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhang, Y., Ashok, S., Seol, E. et al. NADH-dependent lactate dehydrogenase from Alcaligenes eutrophus H16 reduces 2-oxoadipate to 2-hydroxyadipate. Biotechnol Bioproc E 19, 1048–1057 (2014). https://doi.org/10.1007/s12257-014-0381-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-014-0381-1