Abstract
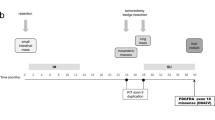
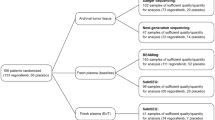
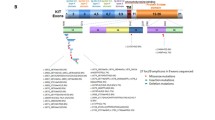
The aim of this study was to characterize secondary kinase mutations in Chinese patients with imatinib-resistant gastrointestinal stromal tumors (GISTs). Mutations in receptor tyrosine kinase (KIT; exons 9, 11, 13, 14, 17, and 18) and platelet-derived growth factor-alpha (PDGFRA; exons 12, 14, and 18) were analyzed by direct sequencing. After imatinib treatment, 425 cancer-related target genes were analyzed by next generation sequencing (NGS) in imatinib-resistant patients. Correlation of sequencing results with clinicopathologic features were analyzed. We identified 320 patients with secondary acquired resistance. We determined that 65.63% (210/320) of resistant patients had secondary KIT mutations in exon 13 (n = 134), exon 14 (n = 10), or exon 17 (n = 66), and 4.38% (14/320) had additional PDGFRA mutations in exon 14 (n = 3) or exon 18 (n = 11). All secondary KIT mutations were missense mutations and were mostly located in kinase domains. Ninety-six imatinib-resistant GIST patients did not have secondary KIT or PDGFRA mutations. Common independent mutation events were found in retinoblastoma protein 1 (RB1) (18/96 cases), SWI/SNF-related matrix associated actin-dependent regulator of chromatin subfamily B member 1 (SMARCB1) (16/96 cases), and myc-associated factor X (MAX) (10/96 cases). RB1 or SMARCB1 mutations coexisted with activation of other oncogenes in 6 or 15 cases, respectively. Multiple mutations were also seen in cases with MAX mutations. These mutations are frequently associated with clinicopathological factors. Secondary mutations of KIT/PDGFRA were the most important contributors in GISTs developing resistance to imatinib treatment. Additional genetic events including RB1, SMARCB1, and MAX except secondary KIT/PDGFRA mutations are the most common for GISTs to evolve into resistant disease. Clinical assessment of the effect of these mutations may benefit existing risk assessment models and selection of adjuvant therapies in GIST patients.




Similar content being viewed by others
References
Di Vita M, Zanghì A, Cavallaro A et al (2019) Gastric GIST and prognostic models. Which is the best to predict survival after surgery? Ann Ital Chir 90:31–40
Kalfusová A, Kodet R (2017) Molecular mechanisms of primary and secondary resistance, molecular-genetic features and characteristics of KIT/PDGFRA non-mutated GISTs. Cesk Patol 53(4):167–173
Miettinen M, Monihan JM, Sarlomo-Rikala M et al (1999) Gastrointestinal stromal tumors/smooth muscle tumors (GISTs) primary in the omentum and mesentery: clinicopathologic and immunohistochemical study of 26 cases. Am J Surg Pathol 23(9):1109–1118
Li J (2016) Molecular mechanism of gastrointestinal stromal tumors and progress in drug research. Zhonghua Wei Chang Wai Ke Za Zhi 19(11):1316–1320
Corless CL, Schroeder A, Griffith D et al (2005) PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol 23(23):5357–5364
Lasota J, Miettinen M (2006) KIT and PDGFRA mutations in gastrointestinal stromal tumors (GISTs). Semin Diagn Pathol 23(2):91–102
Li J, Zhang H, Lu Y et al (2015) Presence of PDGFRA and DOG1 mutations in gastrointestinal stromal tumors among Chinese population. Int J Clin Exp Pathol 8(5):5721–5726
Ahmad F, Lad P, Bhatia S et al (2015) Molecular spectrum of c-KIT and PDGFRA gene mutations in gastro intestinal stromal tumor: determination of frequency, distribution pattern and identification of novel mutations in Indian patients. Med Oncol 32(1):424
Rossi G, Valli R, Bertolini F et al (2005) PDGFR expression in differential diagnosis between KIT-negative gastrointestinal stromal tumours and other primary soft-tissue tumours of the gastrointestinal tract. Histopathology. 46(5):522–531
Heinrich MC, Corless CL, Duensing A et al (2003) PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 299(5607):708–710
Kang HJ, Nam SW, Kim H et al (2005) Correlation of KIT and platelet-derived growth factor receptor alpha mutations with gene activation and expression profiles in gastrointestinal stromal tumors. Oncogene. 24(6):1066–1074
von Mehren M, Joensuu H (2018) Gastrointestinal stromal tumors. J Clin Oncol 36(2):136–143
Saito S, Nakata K, Kajiura S et al (2013) Long-term follow-up outcome of imatinib mesylate treatment for recurrent and unresectable gastrointestinal stromal tumors. Digestion 87(1):47–52
Rodriquenz MG, Rossi S, Ricci R et al (2016) Gastrointestinal stromal tumors (GISTs) and second malignancies: a novel “sentinel tumor”? A monoinstitutional, STROBE-compliant observational analysis. Medicine (Baltimore) 95(38):e4718
Comandone A, Boglione A (2015) The importance of mutational status in prognosis and therapy of GIST. Recenti Prog Med 106(1):17–22
Huang WK, Akçakaya P, Gangaev A et al (2018) miR-125a-5p regulation increases phosphorylation of FAK that contributes to imatinib resistance in gastrointestinal stromal tumors. Exp Cell Res 371(1):287–296
Boyar MS, Taub RN (2007) New strategies for treating GIST when imatinib fails. Cancer Investig 25(5):328–335
Atay S, Wilkey DW, Milhem M et al (2018) Insights into the proteome of gastrointestinal stromal tumors-derived exosomes reveals new potential diagnostic biomarkers. Mol Cell Proteomics 17(3):495–515
Serrano C, Mariño-Enríquez A, Tao DL et al (2019) Complementary activity of tyrosine kinase inhibitors against secondary kit mutations in imatinib-resistant gastrointestinal stromal tumours. Br J Cancer 120(6):612–620
Tsai HJ, Jiaang WT, Shih NY et al (2018) BPR1J373, a novel multitargeted kinase inhibitor, effectively suppresses the growth of gastrointestinal stromal tumor. Cancer Sci 109(11):3591–3601
Li J, Ye Y, Wang J et al (2017) Chinese consensus guidelines for diagnosis and management of gastrointestinal stromal tumor. Chin J Cancer Res 29(4):281–293
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120
Li H, Durbin R (2009) Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 25(14):1754–1760
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNAsequencing data. Genome Res 20(9):1297–1303
Van der Auwera GA, Carneiro MO, Hartl C et al (2013) From FastQ data to high confidence variant calls: the genome analysis toolkit best practicespipeline. Curr Protoc Bioinformatics 43:11.10.1–11.1033
Koboldt DC, Zhang Q, Larson DE et al (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exomesequencing. Genome Res 22(3):568–576
Robinson JT, Thorvaldsdóttir H, Wenger AM et al (2017) Variant review with the integrative genomics viewer. Cancer Res 77(21):e31–e34
Amarasinghe KC, Li J, Hunter SM et al (2014) Inferring copy number and genotype in tumour exome data. BMC Genomics 15:732
Chen W, Kuang Y, Qiu HB et al (2017) Dual targeting of insulin receptor and KIT in Imatinib-resistant gastrointestinal stromal tumors. Cancer Res 77(18):5107–5117
Obata Y, Horikawa K, Shiina I et al (2018) Oncogenic Kit signalling on the Golgi is suppressed by blocking secretory trafficking with M-COPA in gastrointestinal stromal tumours. Cancer Lett 415:1–10
Boichuk S, Galembikova A, Dunaev P et al (2017) A novel receptor tyrosine kinase switch promotes gastrointestinal stromal tumor drug resistance. Molecules. 5:22(12)
Heinrich MC, Maki RG, Corless CL et al (2008) Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J Clin Oncol 26(33):5352–5359
Liegl B, Kepten I, Le C et al (2008) Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J Pathol 216(1):64–74
Yu K, Liu X, Jiang Z et al (2017) Discovery of a highly selective KIT kinase primary V559D mutant inhibitor for gastrointestinal stromal tumors (GISTs). Oncotarget. 8(67):111110–111118
Tamborini E, Pricl S, Negri T et al (2006) Functional analyses and molecular modeling of two c-Kit mutations responsible for imatinib secondary resistance in GIST patients. Oncogene. 25(45):6140–6146
Guo T, Agaram NP, Wong GC et al (2007) Sorafenib inhibits the imatinib-resistant KITT670I gatekeeper mutation in gastrointestinal stromal tumor. Clin Cancer Res 13(16):4874–4881
Antonescu CR, Besmer P, Guo T et al (2005) Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res 11:4182–4190
Lau J, Zhou Q, Sutton SE et al (2014) Inhibition of c-Kit is not required for reversal of hyperglycemia by imatinib in NOD mice. PLoS One 9(1):e84900
Pricl S, Fermeglia M, Ferrone M et al (2005) T315I-mutated Bcr-Abl in chronic myeloid leukemia and imatinib: insights from a computational study. Mol Cancer Ther 4(8):1167–1174
Tutone M, Lauria A, Almerico AM (2011) Study of the role of “gatekeeper” mutations V654A and T670I of c-kit kinase in the interaction withinhibitors by means mixed molecular dynamics/docking approach. Bioinformation. 7(6):296–298
Ihle MA, Huss S, Jeske W et al (2018) Expression of cell cycle regulators and frequency of TP53 mutations in high risk gastrointestinal stromal tumors prior to adjuvant imatinib treatment. PLoS One 13(2):e0193048
Haller F, Gunawan B, von Heydebreck A et al (2005) Prognostic role of E2F1 and members of the CDKN2A network in gastrointestinal stromal tumors. Clin Cancer Res 11(18):6589–6597
Wang X, Lee RS, Alver BH et al (2017) SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 49(2):289–295
Lu C, Allis CD (2017) SWI/SNF complex in cancer. Nat Genet 49(2):178–179
Nakayama RT, Pulice JL, Valencia AM et al (2017) SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet 49(11):1613–1623
Kohashi K, Yamamoto H, Yamada Y et al (2018) SWI/SNF chromatin-remodeling complex status in SMARCB1/INI1-preserved epithelioid sarcoma. Am J Surg Pathol 42(3):312–318
Smith MJ, Walker JA, Shen Y et al (2012) Expression of SMARCB1 (INI1) mutations in familial schwannomatosis. Hum Mol Genet 21(24):5239–5245
Beliakova-Bethell N, Mukim A, White CH et al (2019) Histone deacetylase inhibitors induce complex host responses that contribute to differential potencies of these compounds in HIV reactivation. J Biol Chem 294(14):5576–5589
Mora-Blanco EL, Mishina Y, Tillman EJ et al (2014) Activation of β-catenin/TCF targets following loss of the tumor suppressor SNF5. Oncogene. 33(7):933–938
Kim KH, Roberts CW (2014) Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth. Cancer Genetics 207(9):365–372
Tang CM, Lee TE, Syed SA et al (2016) Hedgehog pathway dysregulation contributes to the pathogenesis of human gastrointestinal stromal tumors via GLI-mediated activation of KIT expression. Oncotarget. 7(48):78226–78241
Lo WW, Wunder JS, Dickson BC et al (2014) Involvement and targeted intervention of dysregulated Hedgehog signaling in osteosarcoma. Cancer. 120(4):537–547
Szczepny A, Rogers S, Jayasekara WSN et al (2017) The role of canonical and non-canonical Hedgehog signaling in tumor progression in a mouse model of small cell lung cancer. Oncogene. 36(39):5544–5550
Pelczar P, Zibat A, van Dop WA et al (2013) Inactivation of Patched1 in mice leads to development of gastrointestinal stromal-like tumors that express Pdgfrα but not kit. Gastroenterology 144(1):134–144. e6
Castell A, Yan Q, Fawkner K et al (2018) A selective high affinity MYC-binding compound inhibits MYC:MAX interaction and MYC-dependent tumor cell proliferation. Sci Rep 8(1)
Melnik S, Werth N, Boeuf S et al (2019) Impact of c-MYC expression on proliferation, differentiation, and risk of neoplastic transformation of human mesenchymal stromal cells. Stem Cell Res Ther 10(1):73
Boyd SR, Young DW (2019) Max-imizing the Attenuation of Myc Using Small Molecules. Trends Pharmacol Sci 40(9):608–612
Struntz NB, Chen A, Deutzmann A et al (2019) Stabilization of the Max Homodimer with a Small Molecule Attenuates Myc-Driven Transcription. Cell Chem Biol 26(5):711–723.e14
Acknowledgements
We thank all patients, physicians, clinical trial nurses, laboratory staff, and technicians for their participation in this study.
Funding
This study was funded by the Natural Science Foundation of the Liaoning province [grant number 20180530045].
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: Jiang Du, Yang Liu. Performed the experiments: Si Wang, Rui Wang and Si-Yao Wang. Analyzed the data: Qiang Han, Hong-Tao Xu and Peng Yang. Contributed reagents/materials/ analysis tools: Jiang Du, Hong-Tao Xu, and Yang Liu. Wrote the paper: All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Ethics Approval and Informed Consent
This study was approved by the Ethics committee of the First Affiliated Hospital of China Medical University and was performed according to the principles of the Declaration of Helsinki. Written informed consents were obtained from the patients in this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Du, J., Wang, S., Wang, R. et al. Identifying Secondary Mutations in Chinese Patients with Imatinib-Resistant Gastrointestinal Stromal Tumors (GISTs) by Next Generation Sequencing (NGS). Pathol. Oncol. Res. 26, 91–100 (2020). https://doi.org/10.1007/s12253-019-00770-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-019-00770-6