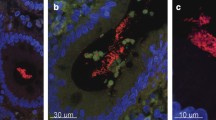
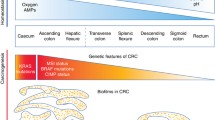
Abstract
Colorectal cancer (CRC) is a heterogeneous disease of the intestinal epithelium and ranks the third largest diagnosed malignancy in the world. Many studies have shown that the high risk of CRC is believed to be related to the formation of biofilms. To prove causation, it will be significant to decipher which specific bacteria in biofilms initiate and maintain CRC and fully describe their underlying mechanisms. Here we introduce a bacterial driver–passenger model. This model added a novel and compelling angle to the role of microorganisms, putting more emphasis on the transformation of bacterial composition in biofilms which play different roles in the development of CRC. In this model, bacterial drivers can initiate the formation of CRC through genotoxicity, while bacterial passengers maintain the CRC process through metabolites. On the basis of these pathogens, we further turned our attention to strategies that can inhibit and eradicate these pathogenic biofilms, with the aim of finding new ways to hinder colorectal carcinogenesis.



Similar content being viewed by others
Data availability
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
References
Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16(12):713–32.
Globocan. https://gco.iarc.fr/today/home Accessed 3 March 2021.
Dai Z, Zhang J, Wu Q, Chen J, Liu J, Wang L, et al. The role of microbiota in the development of colorectal cancer. Int J Cancer. 2019;145(8):2032–41.
Li S, Konstantinov SR, Smits R, Peppelenbosch MP. Bacterial biofilms in colorectal cancer initiation and progression. Trends Mol Med. 2017;23(1):18–30.
Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375):592–7.
Tytgat H, Nobrega F, van der Oost J, Vos de Tim WJ. Bowel biofilms: tipping points between a healthy and compromised gut? Trends Microbiol. 2019;27(1):17–25.
Matsuo K, Ota H, Akamatsu T, Sugiyama A, Katsuyama TJG. Histochemistry of the surface mucous gel layer of the human colon. Trends Microbiol. 1997;40(6):782–9.
Sicard J, Le Bihan G, Vogeleer P, Jacques M, Harel J. Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol. 2017;7:387.
Lappin-Scott H, Costerton JW. Bacterial biofilms and surface fouling. Biofouling. 1989;1(4):323–42.
Roy R, Tiwari M, Donelli G, Tiwari V. Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence. 2018;9(1):522–54.
Stoodley P, Sauer K, Davies D, Costerton JJ. Biofilms as complex differentiated communities. Ann Rev Microbiol. 2002;56:187–209.
Dalton HM, Goodman AE, Marshall KC. Diversity in surface colonization behavior in marine bacteria. J Ind Microbiol. 1996;17(3):228–34.
O’Toole GA, Kolter RJ. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998. https://doi.org/10.1046/j.1365-2958.1998.01062.x.
Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–407.
Tolker-Nielsen T, Brinch UC, Ragas PC, Andersen JB, Jacobsen CS, Molin S. Development and dynamics of Pseudomonas sp. biofilms. J Bacteriol. 2000;182(22):6482–9.
Drewes JL, White JR, Dejea CM, Fathi P, Iyadorai T, Vadivelu J, et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microb. 2017;3:34.
Dejea CM, Wick EC, Hechenbleikner EM, White JR, Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M, Stein E. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Nat Acad Sci. 2014;111(51):18321–6.
Raskov H, Kragh KN, Bjarnsholt T, Alamili M, Gogenur I. Bacterial biofilm formation inside colonic crypts may accelerate colorectal carcinogenesis. Clin Transl Med. 2018;7(1):30.
Soler A, Miller R, Laughlin K, Carp N, Klurfeld D, Mullin J. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis. 1999;20(8):1425–31.
Overhage J, Campisano A, Bains M, Torfs EC, Rehm BH, Hancock RE. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect Immun. 2008;76(9):4176–82.
Ciofu O, Mandsberg LF, Wang H, Høiby N. Phenotypes selected during chronic lung infection in cystic fibrosis patients: implications for the treatment of Pseudomonas aeruginosa biofilm infections. FEMS Immunol Med Microbiol. 2012;65(2):215–25.
Anderl JN, Zahller J, Roe F, Stewart PS. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2003;47(4):1251–6.
Walters MC 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47(1):317–23.
Ma H, Bryers JD. Non-invasive determination of conjugative transfer of plasmids bearing antibiotic-resistance genes in biofilm-bound bacteria: effects of substrate loading and antibiotic selection. Appl Microbiol Biotechnol. 2013;97(1):317–28.
Ibrahim NH, Melake NA, Somily AM, Zakaria AS, Baddour MM, Mahmoud AZ. The effect of antifungal combination on transcripts of a subset of drug-resistance genes in clinical isolates of Candida species induced biofilms. Saudi Pharm J. 2015;23(1):55–66.
Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21(6):891–7.
Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10(8):575–82.
Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, et al. Towards the human colorectal cancer microbiome. PLoS ONE. 2011;6(5):e20447.
Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507.
Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306.
Harald zur H. The search for infectious causes of human cancers: where and why. Virology. 2009;392(1):1.
Boleij A, Tjalsma H. Gut bacteria in health and disease: a survey on the interface between intestinal microbiology and colorectal cancer. Biol Rev Camb Philos Soc. 2012;87(3):701–30.
Wu S, Shin J, Zhang G, Cohen M, Franco A, Sears CL. The Bacteroides fragilis toxin binds to a specific intestinal epithelial cell receptor. Infect Immun. 2006;74(9):5382–90.
Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12(8):782–6.
Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci USA. 2011;108(37):15354–9.
Abais JM, Xia M, Zhang Y, Boini KM, Li PL. Redox regulation of NLRP3 inflammasomes: ROS as trigger or effector? Antioxid Redox Signal. 2015;22(13):1111–29.
Wu S, Rhee KJ, Zhang M, Franco A, Sears CL. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J Cell Sci. 2007;120(Pt 11):1944–52.
Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361(25):2449–60.
Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124(2):392–400.
Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–22.
Göktuna SI, Shostak K, Chau TL, Heukamp LC, Hennuy B, Duong HQ, et al. The prosurvival IKK-related kinase IKKε integrates LPS and IL17A signaling cascades to promote Wnt-dependent tumor development in the intestine. Can Res. 2016;76(9):2587–99.
Hata K, Andoh A, Shimada M, Fujino S, Bamba S, Araki Y, et al. IL-17 stimulates inflammatory responses via NF-kappaB and MAP kinase pathways in human colonic myofibroblasts. Am J Physiol Gastrointest Liver Physiol. 2002;282(6):G1035–44.
Swidsinski A, Weber J, Loening-Baucke V, Hale L, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43(7):3380–9.
Ruiz PA, Shkoda A, Kim SC, Sartor RB, Haller D. IL-10 gene-deficient mice lack TGF-beta/Smad-mediated TLR2 degradation and fail to inhibit proinflammatory gene expression in intestinal epithelial cells under conditions of chronic inflammation. Ann N Y Acad Sci. 2006;1072:389–94.
Szigeti R, Pangas SA, Nagy-Szakal D, Dowd SE, Shulman RJ, Olive AP, et al. SMAD4 haploinsufficiency associates with augmented colonic inflammation in select humans and mice. Ann Clin Lab Sci. 2012;42(4):401–8.
Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 2007;132(2):551–61.
McCool KW, Miyamoto S. DNA damage-dependent NF-κB activation: NEMO turns nuclear signaling inside out. Immunol Rev. 2012;246(1):311–26.
Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer. 2001;1(2):109–17.
Zhou H, Ivanov VN, Gillespie J, Geard CR, Amundson SA, Brenner DJ, et al. Mechanism of radiation-induced bystander effect: role of the cyclooxygenase-2 signaling pathway. Proc Natl Acad Sci USA. 2005;102(41):14641–6.
Carniol K, Gilmore MS. Signal transduction, quorum-sensing, and extracellular protease activity in Enterococcus faecalis biofilm formation. J Bacteriol. 2004;186(24):8161–3.
Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science (New York, NY). 2006;313(5788):848–51.
Allen-Vercoe E, Jobin C. Fusobacterium and Enterobacteriaceae: important players for CRC. Immunol Lett. 2014;162:54–61.
Leung A, Tsoi H, Yu J. Fusobacterium and Escherichia: models of colorectal cancer driven by microbiota and the utility of microbiota in colorectal cancer screening. Expert Rev Gastroenterol Hepatol. 2015;9(5):651–7.
Maddocks OD, Scanlon KM, Donnenberg MS. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. mBio. 2013;4(3):e00152-e213.
Housseau F, Sears CL. Enterotoxigenic Bacteroides fragilis (ETBF)-mediated colitis in Min (Apc+/-) mice: a human commensal-based murine model of colon carcinogenesis. Cell cycle (Georgetown, Tex). 2010;9(1):3–5.
Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microb. 2010;1(3):138–47.
Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Can Res. 2009;69(11):4918–25.
Cao H, Luo S, Xu M, Zhang Y, Song S, Wang S, et al. The secondary bile acid, deoxycholate accelerates intestinal adenoma-adenocarcinoma sequence in Apc (min/+) mice through enhancing Wnt signaling. Fam Cancer. 2014;13(4):563–71.
Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158(2):288–99.
den Besten G, Lange K, Havinga R, van Dijk TH, Gerding A, van Eunen K, et al. Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am J Physiol Gastrointest Liver Physiol. 2013;305(12):G900–10.
Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. 2014;111(6):2247–52.
Kim CH, Park J, Kim M. Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation. Immun Netw. 2014;14(6):277–88.
Heerdt BG, Houston MA, Augenlicht LH. Potentiation by specific short-chain fatty acids of differentiation and apoptosis in human colonic carcinoma cell lines. Can Res. 1994;54(12):3288–93.
Biarc J, Nguyen IS, Pini A, Gossé F, Richert S, Thiersé D, et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S. bovis). Carcinogenesis. 2004;25(8):1477–84.
Abdulamir AS, Hafidh RR, Abu BF. The association of Streptococcus bovis/gallolyticus with colorectal tumors: the nature and the underlying mechanisms of its etiological role. J Exp Clin Cancer Res CR. 2011;30(1):11.
Ohshima H, Bartsch H. Chronic infections and inflammatory processes as cancer risk factors: possible role of nitric oxide in carcinogenesis. Mutat Res. 1994;305(2):253–64.
Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer. 2010;9:249.
Deng Q, Wang C, Yu K, Wang Y, Yang Q, Zhang J, et al. Streptococcus bovis contributes to the development of colorectal cancer via recruiting CD11b+ TLR-4+ Cells. Med Sci Monit. 2020;26:e921886.
Boleij A, van Gelder MM, Swinkels DW, Tjalsma H. Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis. 2011;53(9):870–8.
Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2011;55(9):4469–74.
Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. In vivo pharmacokinetics/pharmacodynamics of colistin and imipenem in Pseudomonas aeruginosa biofilm infection. Antimicrob Agents Chemother. 2012;56(5):2683–90.
Hussain A, Alajmi MF, Khan MA, Pervez SA, Ahmed F, Amir S, et al. Biosynthesized Silver Nanoparticle (AgNP) from pandanus odorifer leaf extract exhibits anti-metastasis and anti-biofilm potentials. Front Microbiol. 2019;10:8.
Gopinath K, Kumaraguru S, Bhakyaraj K, Mohan S, Venkatesh KS, Esakkirajan M, et al. Green synthesis of silver, gold and silver/gold bimetallic nanoparticles using the Gloriosa superba leaf extract and their antibacterial and antibiofilm activities. Microb Pathog. 2016;101:1–11.
Singh R, Dumlupinar G, Andersson-Engels S, Melgar S. Emerging applications of upconverting nanoparticles in intestinal infection and colorectal cancer. Int J Nanomed. 2019;14:1027–38.
Nayak M, Singh AK, Prakash P, Kant R, Bhattacharya S. Structural studies on thiosalicylate complexes of Zn (II) & Hg (II). First insight into Zn (II)-thiosalicylate complex as potential antibacterial, antibiofilm and anti-tumour agent. Inorg Chim Acta. 2020;501:119263.
Jang HI, Eom YB. Antibiofilm and antibacterial activities of repurposing auranofin against Bacteroides fragilis. Arch Microbiol. 2020. https://doi.org/10.1007/s00203-019-01764-3.
Sambanthamoorthy K, Luo C, Pattabiraman N, Feng X, Koestler B, Waters CM, et al. Identification of small molecules inhibiting diguanylate cyclases to control bacterial biofilm development. Biofouling. 2014;30(1–2):17–28.
Kim JY, Park SC, Yoon MY, et al. C-terminal amidation of PMAP-23: translocation to the inner membrane of Gram-negative bacteria. Amino Acids. 2011. https://doi.org/10.1007/s00726-010-0632-1.
Mogi T, Kita K. Gramicidin S and polymyxins: the revival of cationic cyclic peptide antibiotics. Cell Mol Life Sci. 2009;66(23):3821–6.
Kragol G, Hoffmann R, Chattergoon MA, Lovas S, Cudic M, Bulet P, et al. Identification of crucial residues for the antibacterial activity of the proline-rich peptide, pyrrhocoricin. Eur J Biochem. 2002. https://doi.org/10.1046/j.1432-1033.2002.03119.x.
Vizán J, Hernandez-Chico CI, Castillo ID, Moreno F. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. The EMBO journal. 1999;10(2):467–76.
Kharidia R, Liang J. The activity of a small lytic peptide PTP-7 on Staphylococcus aureus biofilms. J Microbiol. 2011;49(4):663–8.
Overhage J, Campisano A, Bains M, Torfs E, Hancock R. Immunity human host defense peptide LL-37 prevents bacterial biofilm formation. Infect Immun. 2008;76(9):4176–82.
Boman HG. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect Immun. 1993;61(7):2978–84.
Hsu CH, Chen C, Jou ML, Lee YL, Wu SH. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 2005;33(13):4053–64.
Ju HC, Sung BH, Sun CK. Buforins: histone H2A-derived antimicrobial peptides from toad stomach. Biochim BiophysActa Biomembr. 2009;1788(8):1564–9.
Ma DSL, Tan LT, Chan KG, Yap WH, Pusparajah P, Chuah LH, et al. Resveratrol-potential antibacterial agent against foodborne pathogens. Front Pharmacol. 2018;9:102.
Santosh Kumar SC, Srinivas P, Negi PS, Bettadaiah BK. Antibacterial and antimutagenic activities of novel zerumbone analogues. Food Chem. 2013;141(2):1097–103.
Haque MA, Jantan I, Arshad L, Bukhari SNA. Exploring the immunomodulatory and anticancer properties of zerumbone. Food Funct. 2017;8(10):3410–31.
Kim HR, Rhee KJ, Eom YB. Anti-biofilm and antimicrobial effects of zerumbone against Bacteroides fragilis. Anaerobe. 2019;57:99–106.
Jang HI, Rhee KJ, Eom YB. Antibacterial and antibiofilm effects of α-humulene against Bacteroides fragilis. Can J Microbiol. 2020;66(6):389–99.
Chmit M, Kanaan H, Habib J, Abbass M, McHeik A, Chokr A. Antibacterial and antibiofilm activities of polysaccharides, essential oil, and fatty oil extracted from Laurus nobilis growing in Lebanon. Asian Pac J Trop Med. 2014;71:546–52.
Bakkiyaraj D, Nandhini JR, Malathy B, Pandian SK. The anti-biofilm potential of pomegranate (Punica granatum L.) extract against human bacterial and fungal pathogens. Biofouling. 2013;29(8):929–37.
Tan LT, Chan KG, Lee LH, Goh BH. Streptomyces bacteria as potential probiotics in aquaculture. Front Microbiol. 2016;7:79.
Shin DS, Rhee KJ, Eom YB. Effect of probiotic clostridium butyricum NCTC 7423 supernatant on biofilm formation and gene expression of Bacteroides fragilis. J Microbiol Biotechnol. 2020;30(3):368–77.
Abdelhamid AG, Esaam A, Hazaa MM. Cell free preparations of probiotics exerted antibacterial and antibiofilm activities against multidrug resistant E. coli. Saudi Pharm J. 2018;26(5):603–7.
Mukai T, Kaneko S, Matsumoto M, Ohori H. Binding of Bifidobacterium bifidum and Lactobacillus reuteri to the carbohydrate moieties of intestinal glycolipids recognized by peanut agglutinin. Int J Food Microbiol. 2004;90(3):357–62.
Barzegari A, Kheyrolahzadeh K, Hosseiniyan Khatibi SM, Sharifi S, Memar MY, Zununi VS. The battle of probiotics and their derivatives against biofilms. Infect Drug Res. 2020;13:659–72.
Mahdhi A, Leban N, Chakroun I, Bayar S, Mahdouani K, Majdoub H, et al. Use of extracellular polysaccharides, secreted by Lactobacillus plantarum and Bacillus, as reducing indole production agents to control biofilm formation and efflux pumps inhibitor in Escherichia coli. Microb Pathog. 2018;125:448–53.
Sharma V, Harjai K, Shukla G. Effect of bacteriocin and exopolysaccharides isolated from probiotic on P aeruginosa PAO1 biofilm. Folia Microbiol. 2018;63(2):181–90.
Jiang Q, Chen J, Yang C, Yin Y, Yao K. Quorum sensing: a prospective therapeutic target for bacterial diseases. Biomed Res Int. 2019;2019:2015978.
Zhao X, Yu Z, Ding T. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms. 2020. https://doi.org/10.3390/microorganisms8030425.
Walz JM, Avelar RL, Longtine KJ, Carter KL, Mermel LA, Heard SO. Anti-infective external coating of central venous catheters: a randomized, noninferiority trial comparing 5-fluorouracil with chlorhexidine/silver sulfadiazine in preventing catheter colonization. Crit Care Med. 2010;38(11):2095–102.
van Delden C, Köhler T, Brunner-Ferber F, François B, Carlet J, Pechère JC. Azithromycin to prevent Pseudomonas aeruginosa ventilator-associated pneumonia by inhibition of quorum sensing: a randomized controlled trial. Intensive Care Med. 2012;38(7):1118–25.
Funding
This study was supported by Training Project of Key Talents of Youth Medicine in Jiangsu province, China [No. QNRC2016330], The Key disease standardization diagnosis and treatment project in Jiangsu province [BE2015664], The Academic Science and Technology Innovation Fund for College Students [No. X20180714], the Social Development-Health Care Project of Yangzhou, Jiangsu Province [No. YZ2018087], the Social Development Project of Yangzhou, Jiangsu Province [No. YZ2021075], 2021 Jiangsu Graduate Research And Innovation Program (SJCX21 1644) and High-level talent “six one project” top talent scientific project of Jiangsu Province [No. LGY2019034].
Author information
Authors and Affiliations
Contributions
JX and YF drafted the manuscript in detail. JX and HZ researched the literatures and drawn figures. YF, YC and WZ plotted the diagram and table. DT and DW critically revised the article for important intellectual content. JX and YF have contributed equally to this work and share first authorship. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
All authors have given their consent for their participation.
Consent for publication
All authors have given their consent for the publishing of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xing, J., Fang, Y., Zhang, W. et al. Bacterial driver–passenger model in biofilms: a new mechanism in the development of colorectal cancer. Clin Transl Oncol 24, 784–795 (2022). https://doi.org/10.1007/s12094-021-02738-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-021-02738-y