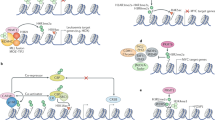
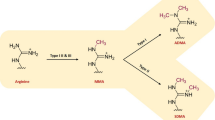
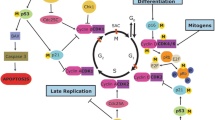
Abstract
A remarkable post-transitional modification of both histones and non-histone proteins is arginine methylation. Methylation of arginine residues is crucial for a wide range of cellular process, including signal transduction, DNA repair, gene expression, mRNA splicing, and protein interaction. Arginine methylation is modulated by arginine methyltransferases and demethylases, like protein arginine methyltransferase (PRMTs) and Jumonji C (JmjC) domain containing (JMJD) proteins. Symmetric dimethylarginine and asymmetric dimethylarginine, metabolic products of the PRMTs and JMJD proteins, can be changed by abnormal expression of these proteins. Many pathologies including cancer, inflammation and immune responses have been closely linked to aberrant arginine methylation. Currently, the majority of the literature discusses the substrate specificity and function of arginine methylation in the pathogenesis and prognosis of cancers. Numerous investigations on the roles of arginine methylation in the central nervous system (CNS) have so far been conducted. In this review, we display the biochemistry of arginine methylation and provide an overview of the regulatory mechanism of arginine methyltransferases and demethylases. We also highlight physiological functions of arginine methylation in the CNS and the significance of arginine methylation in a variety of neurological diseases such as brain cancers, neurodegenerative diseases and neurodevelopmental disorders. Furthermore, we summarize PRMT inhibitors and molecular functions of arginine methylation. Finally, we pose important questions that require further research to comprehend the roles of arginine methylation in the CNS and discover more effective targets for the treatment of neurological diseases.






Similar content being viewed by others
Data Availability
Not applicable.
Abbreviations
- 7SK RNA:
-
7SK small nuclear RNA
- Aβ:
-
Amyloid-beta
- Acsl4:
-
Acyl-CoA synthetase long chain family member 4
- AD:
-
Alzheimer’ s disease
- ADMA:
-
Asymmetrical dimethylarginine
- AIF:
-
Apoptosis-inducing factor
- ALDH1A1:
-
Aldehyde dehydrogenase 1
- ALS:
-
Amyotrophic lateral sclerosis
- APP:
-
Amyloid precursor protein
- AR:
-
Androgen receptor
- ATF5:
-
Activating transcription factor 5
- Btg2:
-
B-cell translocation gene 2
- CCI:
-
Chronic constriction injury
- CDKN2A:
-
Cyclin-dependent kinase inhibitor 2A
- CK2:
-
Casein kinase 2
- CNS:
-
Central nervous system
- Cobl:
-
Cordon-bleu WH2 repeat protein
- COPI:
-
Coat protein complex I
- CPP:
-
Conditioned place preference
- CREB1:
-
CAMP response-element-binding protein 1
- DIs:
-
Detained introns
- E2F2:
-
E2F transcription factor 2
- ER:
-
Estrogen receptor
- ESCs:
-
Embryonic stem cells
- FBXO10:
-
F-box protein O10
- FOXP1:
-
Forkhead box protein P1
- FUS:
-
Fused in sarcoma/translocated in liposarcoma
- GSK3β:
-
Glycogen synthase kinase 3β
- GAR:
-
Glycine/arginine-rich
- GBM:
-
Glioblastoma
- GBMNS:
-
Glioblastoma neurospheres
- GSCs:
-
Glioblastoma stem cells
- H3R8:
-
Histone H3 arginine-8
- H3R8me2a:
-
H3R8 asymmetric methylation
- HCN:
-
Hyperpolarization-activated cyclic nucleotide-gated
- HD:
-
Huntington's disease
- HEK:
-
Human embryonic kidney
- HIF-1α:
-
Hypoxia-inducible factor-1 alpha
- HNK-1:
-
Human natural killer-1
- hnRNPA2:
-
Heterogeneous nuclear ribonucleoprotein A2
- HOXC10:
-
Homeobox C10
- HSP70:
-
Heat shock protein 70
- HTT:
-
Huntingtin
- INEA:
-
Injectable n-butyl-2-cyanoacrylate ethyl oleate implant
- IRES:
-
Internal ribosome entry site
- JMJD6:
-
Jumonji C (JmjC) domain containing protein 6
- KCNQ:
-
Potassium voltage-gated channel subfamily KQT
- KSRP:
-
KH-type splicing regulatory protein
- MECP2:
-
Methyl-CpG binding protein 2
- MMA:
-
Monomethylated arginine
- MLL4:
-
Mixed-lineage leukemia 4
- MPP+ :
-
1-Methyl-4-phenylpyridinium iodide
- MTAP:
-
Methylthioadenosine phosphorylase
- mTOR:
-
Mammalian target of rapamycin
- NAc:
-
Nucleus accumbens
- NALCN:
-
Sodium leak channel
- NaV:
-
Sodium channel
- ncRNAs:
-
Non-coding RNAs
- NDUFAF7:
-
NADH:ubiquinone oxidoreductase complex assembly factor 7
- NDUFS2:
-
NADH dehydrogenase[ubiquinone] iron-sulfur protein 2
- NF-κB:
-
Nuclear factor-κB
- NGF:
-
Nerve growth factor
- NS/PCs:
-
Neural stem/precursor cells
- NSCs:
-
Neural stem cells
- NOS:
-
Nitric oxide synthase
- OGD/R:
-
Oxygen-glucose deprivation/reoxygenation
- PAME:
-
Palmitic acid methyl ester
- PARP1:
-
Poly (ADP-ribose) polymerase-1
- PC:
-
Phosphatidylcholine
- PD:
-
Parkinson’s disease
- PDGFRα:
-
Platelet-derived growth factor receptor alpha
- PI3K:
-
Phosphoinositide 3-kinase
- PLD:
-
Phospholipase D
- PRC1:
-
Polycomb repressive complex 1
- PRMT:
-
Protein arginine methyltransferase
- PTEN:
-
Phosphatase and tensin homolog
- PTMs:
-
Post-translational modifications
- RAGE:
-
Receptor for advanced glycosylation end products
- RALY:
-
RNA-binding protein associated with lethal yellow mutation
- RCC1:
-
Regulator of chromosome condensation 1
- RNF168:
-
Ring finger protein 168
- rpS2:
-
Ribosomal protein S2
- SAM:
-
S-adenosyl-l-methionine
- SAP145:
-
Splicing factor SF3B2
- SBMA:
-
Spinobulbar muscular atrophy
- SC1:
-
Schwann cell factor 1
- SCYL1:
-
SCY1-like pseudokinase 1
- SDMA:
-
Symmetric dimethylarginine
- SER-2:
-
Tyramine receptor
- SH3:
-
Src Homology 3
- SMA:
-
Spinal muscular atrophy
- SMN:
-
Survival motor neuron protein
- SNRP:
-
Small nuclear ribonucleoprotein
- ST7:
-
Suppression of tumorigenicity 7
- TBI:
-
Traumatic brain injury
- TDP43:
-
TAR DNA-binding protein 43
- TNR:
-
Tenascin-R
- TRAF6:
-
Tumor necrosis factor receptor-associated factor 6
- VEGF:
-
Vascular endothelial growth factor
- WDR5:
-
WD-40 repeat-containing protein 5
References
Paik WK, Kim S (1969) Enzymatic methylation of histones. Arch Biochem Biophys 134(2):632–637. https://doi.org/10.1016/0003-9861(69)90327-0
Paik WK, Kim S (1968) Protein methylase I. Purification and properties of the enzyme. J Biol Chem 243(9):2108–2114
Lu SC, Mato JM (2012) S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev 92(4):1515–1542. https://doi.org/10.1152/physrev.00047.2011
Blanc RS, Richard S (2017) Arginine methylation: the coming of age. Mol Cell 65(1):8–24. https://doi.org/10.1016/j.molcel.2016.11.003
Wang K, Yang C, Li H, Liu X, Zheng M, Xuan Z, Mei Z, Wang H (2022) Role of the epigenetic modifier JMJD6 in tumor development and regulation of immune response. Front Immunol 13:859893. https://doi.org/10.3389/fimmu.2022.859893
Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 24(21):9630–9645. https://doi.org/10.1128/mcb.24.21.9630-9645.2004
Yan F, Alinari L, Lustberg ME, Martin LK, Cordero-Nieves HM, Banasavadi-Siddegowda Y, Virk S, Barnholtz-Sloan J et al (2014) Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res 74(6):1752–1765. https://doi.org/10.1158/0008-5472.Can-13-0884
Wong M, Sun Y, Xi Z, Milazzo G, Poulos RC, Bartenhagen C, Bell JL, Mayoh C et al (2019) JMJD6 is a tumorigenic factor and therapeutic target in neuroblastoma. Nat Commun 10(1):3319. https://doi.org/10.1038/s41467-019-11132-w
Dong F, Li Q, Yang C, Huo D, Wang X, Ai C, Kong Y, Sun X et al (2018) PRMT2 links histone H3R8 asymmetric dimethylation to oncogenic activation and tumorigenesis of glioblastoma. Nat Commun 9(1):4552. https://doi.org/10.1038/s41467-018-06968-7
Tradewell ML, Yu Z, Tibshirani M, Boulanger MC, Durham HD, Richard S (2012) Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet 21(1):136–149. https://doi.org/10.1093/hmg/ddr448
Rhein VF, Carroll J, Ding S, Fearnley IM, Walker JE (2013) NDUFAF7 methylates arginine 85 in the NDUFS2 subunit of human complex I. J Biol Chem 288(46):33016–33026. https://doi.org/10.1074/jbc.M113.518803
Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR (2000) PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem 275(11):7723–7730. https://doi.org/10.1074/jbc.275.11.7723
Hadjikyriacou A, Yang Y, Espejo A, Bedford MT, Clarke SG (2015) Unique features of human protein arginine methyltransferase 9 (PRMT9) and its substrate RNA splicing factor SF3B2. J Biol Chem 290(27):16723–16743. https://doi.org/10.1074/jbc.M115.659433
Wu Q, Schapira M, Arrowsmith CH, Barsyte-Lovejoy D (2021) Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat Rev Drug Discov 20(7):509–530. https://doi.org/10.1038/s41573-021-00159-8
Jeong HJ, Lee HJ, Vuong TA, Choi KS, Choi D, Koo SH, Cho SC, Cho H et al (2016) Prmt7 deficiency causes reduced skeletal muscle oxidative metabolism and age-related obesity. Diabetes 65(7):1868–1882. https://doi.org/10.2337/db15-1500
Bouchard C, Sahu P, Meixner M, Nötzold RR, Rust MB, Kremmer E, Feederle R, Hart-Smith G et al (2018) Genomic location of PRMT6-dependent H3R2 methylation is linked to the transcriptional outcome of associated genes. Cell Rep 24(12):3339–3352. https://doi.org/10.1016/j.celrep.2018.08.052
Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT (2005) PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem 280(38):32890–32896. https://doi.org/10.1074/jbc.M506944200
Cockman ME, Sugimoto Y, Pegg HB, Masson N, Salah E, Tumber A, Flynn HR, Kirkpatrick JM et al (2022) Widespread hydroxylation of unstructured lysine-rich protein domains by JMJD6. Proc Natl Acad Sci U S A 119(32):e2201483119. https://doi.org/10.1073/pnas.2201483119
Xiao RQ, Ran T, Huang QX, Hu GS, Fan DM, Yi J, Liu W (2022) A specific JMJD6 inhibitor potently suppresses multiple types of cancers both in vitro and in vivo. Proc Natl Acad Sci U S A 119(34):e2200753119. https://doi.org/10.1073/pnas.2200753119
Li S, Ali S, Duan X, Liu S, Du J, Liu C, Dai H, Zhou M et al (2018) JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep 23(2):389–403. https://doi.org/10.1016/j.celrep.2018.03.051
Frankel A, Clarke S (2000) PRMT3 is a distinct member of the protein arginine N-methyltransferase family: conferral of substrate specificity by a zinc-finger domain. J Biol Chem 275(42):32974–32982. https://doi.org/10.1074/jbc.M006445200
Suresh S, Huard S, Dubois T (2021) CARM1/PRMT4: making its mark beyond its function as a transcriptional coactivator. Trends Cell Biol 31(5):402–417. https://doi.org/10.1016/j.tcb.2020.12.010
Musiani D, Bok J, Massignani E, Wu L, Tabaglio T, Ippolito MR, Cuomo A, Ozbek U et al (2019) Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci Signal 12(575). https://doi.org/10.1126/scisignal.aat8388
Yang Y, Hadjikyriacou A, Xia Z, Gayatri S, Kim D, Zurita-Lopez C, Kelly R, Guo A et al (2015) PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat Commun 6:6428. https://doi.org/10.1038/ncomms7428
Gayatri S, Cowles MW, Vemulapalli V, Cheng D, Sun Z-W, Bedford MT (2016) Using oriented peptide array libraries to evaluate methylarginine-specific antibodies and arginine methyltransferase substrate motifs. Sci Rep 6(1):28718. https://doi.org/10.1038/srep28718
Fulton MD, Cao M, Ho MC, Zhao X, Zheng YG (2021) The macromolecular complexes of histones affect protein arginine methyltransferase activities. J Biol Chem 297(4):101123. https://doi.org/10.1016/j.jbc.2021.101123
Yao B, Gui T, Zeng X, Deng Y, Wang Z, Wang Y, Yang D, Li Q et al (2021) PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling. Genome Med 13(1):58. https://doi.org/10.1186/s13073-021-00871-5
Min Z, Xiaomeng L, Zheng L, Yangge D, Xuejiao L, Longwei L, Xiao Z, Yunsong L et al (2019) Asymmetrical methyltransferase PRMT3 regulates human mesenchymal stem cell osteogenesis via miR-3648. Cell Death Dis 10(8):581. https://doi.org/10.1038/s41419-019-1815-7
Shen Y, Li F, Szewczyk MM, Halabelian L, Chau I, Eram MS, Dela Seña C, Park K-S et al (2021) A first-in-class, highly selective and cell-active allosteric inhibitor of protein arginine methyltransferase 6. J Med Chem 64(7):3697–3706. https://doi.org/10.1021/acs.jmedchem.0c02160
Zhang Y, van Haren MJ, Martin NI (2020) Peptidic transition state analogues as PRMT inhibitors. Methods 175:24–29. https://doi.org/10.1016/j.ymeth.2019.08.003
Smith E, Zhou W, Shindiapina P, Sif S, Li C, Baiocchi RA (2018) Recent advances in targeting protein arginine methyltransferase enzymes in cancer therapy. Expert Opin Ther Targets 22(6):527–545. https://doi.org/10.1080/14728222.2018.1474203
Waldmann T, Izzo A, Kamieniarz K, Richter F, Vogler C, Sarg B, Lindner H, Young NL et al (2011) Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 4:11. https://doi.org/10.1186/1756-8935-4-11
Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD et al (2001) Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol 11(24):1981–1985. https://doi.org/10.1016/S0960-9822(01)00600-5
Casadio F, Lu X, Pollock SB, LeRoy G, Garcia BA, Muir TW, Roeder RG, Allis CD (2013) H3R42me2a is a histone modification with positive transcriptional effects. Proc Natl Acad Sci U S A 110(37):14894–14899. https://doi.org/10.1073/pnas.1312925110
Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR et al (2001) Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 40(19):5747–5756. https://doi.org/10.1021/bi002631b
Schneider L, Herkt S, Wang L, Feld C, Wesely J, Kuvardina ON, Meyer A, Oellerich T et al (2021) PRMT6 activates cyclin D1 expression in conjunction with the transcription factor LEF1. Oncogenesis 10(5):42. https://doi.org/10.1038/s41389-021-00332-z
Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer U-M (2007) PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev 21(24):3369–3380. https://doi.org/10.1101/gad.447007
Migliori V, Müller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J et al (2012) Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol 19(2):136–144. https://doi.org/10.1038/nsmb.2209
Morita K, Hatanaka Y, Ihashi S, Asano M, Miyamoto K, Matsumoto K (2021) Symmetrically dimethylated histone H3R2 promotes global transcription during minor zygotic genome activation in mouse pronuclei. Sci Rep 11(1):10146. https://doi.org/10.1038/s41598-021-89334-w
Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ et al (2009) PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol 16(3):304–311. https://doi.org/10.1038/nsmb.1568
Fulton MD, Brown T, Zheng YG (2018) Mechanisms and inhibitors of histone arginine methylation. Chem Rec 18(12):1792–1807. https://doi.org/10.1002/tcr.201800082
Kim H, Ronai ZA (2020) PRMT5 function and targeting in cancer. Cell Stress 4(8):199–215. https://doi.org/10.15698/cst2020.08.228
Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG (2012) Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. J Biol Chem 287(11):7859–7870. https://doi.org/10.1074/jbc.M111.336271
Srour N, Villarreal OD, Hardikar S, Yu Z, Preston S, Miller WH Jr, Szewczyk MM, Barsyte-Lovejoy D et al (2022) PRMT7 ablation stimulates anti-tumor immunity and sensitizes melanoma to immune checkpoint blockade. Cell Reports 38(13):110582. https://doi.org/10.1016/j.celrep.2022.110582
Karkhanis V, Wang L, Tae S, Hu YJ, Imbalzano AN, Sif S (2012) Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase δ catalytic subunit gene, POLD1. J Biol Chem 287(35):29801–29814. https://doi.org/10.1074/jbc.M112.378281
Jain K, Jin CY, Clarke SG (2017) Epigenetic control via allosteric regulation of mammalian protein arginine methyltransferases. Proc Natl Acad Sci U S A 114(38):10101–10106. https://doi.org/10.1073/pnas.1706978114
Feng Y, Maity R, Whitelegge JP, Hadjikyriacou A, Li Z, Zurita-Lopez C, Al-Hadid Q, Clark AT et al (2013) Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J Biol Chem 288(52):37010–37025. https://doi.org/10.1074/jbc.M113.525345
Chang B, Chen Y, Zhao Y, Bruick RK (2007) JMJD6 is a histone arginine demethylase. Science 318(5849):444–447. https://doi.org/10.1126/science.1145801
Shen C, Quan Q, Yang C, Wen Y, Li H (2018) Histone demethylase JMJD6 regulates cellular migration and proliferation in adipose-derived mesenchymal stem cells. Stem Cell Res Ther 9(1):212. https://doi.org/10.1186/s13287-018-0949-3
Yang Y, Bedford MT (2013) Protein arginine methyltransferases and cancer. Nat Rev Cancer 13(1):37–50. https://doi.org/10.1038/nrc3409
Auclair Y, Richard S (2013) The role of arginine methylation in the DNA damage response. DNA Repair 12(7):459–465. https://doi.org/10.1016/j.dnarep.2013.04.006
Al-Hamashi AA, Diaz K, Huang R (2020) Non-histone arginine methylation by protein arginine methyltransferases. Curr Protein Pept Sci 21(7):699–712. https://doi.org/10.2174/1389203721666200507091952
Manni W, Jianxin X, Weiqi H, Siyuan C, Huashan S (2022) JMJD family proteins in cancer and inflammation. Signal Transduct Target Ther 7(1):304. https://doi.org/10.1038/s41392-022-01145-1
Herrmann F, Lee J, Bedford MT, Fackelmayer FO (2005) Dynamics of human protein arginine methyltransferase 1(PRMT1) in vivo. J Biol Chem 280(45):38005–38010. https://doi.org/10.1074/jbc.M502458200
Hashimoto M, Fukamizu A, Nakagawa T (1865) Kizuka Y (2021) Roles of protein arginine methyltransferase 1 (PRMT1) in brain development and disease. BBA-Gen Subjects 1:129776. https://doi.org/10.1016/j.bbagen.2020.129776
Hou W, Nemitz S, Schopper S, Nielsen ML, Kessels MM, Qualmann B (2018) Arginine methylation by PRMT2 controls the functions of the actin nucleator cobl. Dev Cell 45(2):262-275.e268. https://doi.org/10.1016/j.devcel.2018.03.007
Ikenaka K, Miyata S, Mori Y, Koyama Y, Taneda T, Okuda H, Kousaka A, Tohyama M (2006) Immunohistochemical and western analyses of protein arginine N-methyltransferase 3 in the mouse brain. Neuroscience 141(4):1971–1982. https://doi.org/10.1016/j.neuroscience.2006.05.022
Huang J, Vogel G, Yu Z, Almazan G, Richard S (2011) Type II arginine methyltransferase PRMT5 regulates gene expression of inhibitors of differentiation/DNA binding Id2 and Id4 during glial cell differentiation. J Biol Chem 286:44424–44432. https://doi.org/10.1074/jbc.M111.277046
Lim CS, Alkon DL (2017) Inhibition of coactivator-associated arginine methyltransferase 1 modulates dendritic arborization and spine maturation of cultured hippocampal neurons. J Biol Chem 292(15):6402–6413. https://doi.org/10.1074/jbc.M117.775619
Lee J, Villarreal OD, Wang YC, Ragoussis J, Rivest S, Gosselin D, Richard S (2022) PRMT1 is required for the generation of MHC-associated microglia and remyelination in the central nervous system. Life Sci Alliance 5(10). https://doi.org/10.26508/lsa.202201467
Quan X, Yue W, Luo Y, Cao J, Wang H, Wang Y, Lu Z (2015) The protein arginine methyltransferase PRMT5 regulates Aβ-induced toxicity in human cells and Caenorhabditis elegans models of Alzheimer’s disease. J Neurochem 134(5):969–977. https://doi.org/10.1111/jnc.13191
Chittka A, Nitarska J, Grazini U, Richardson WD (2012) Transcription factor positive regulatory domain 4 (PRDM4) recruits protein arginine methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J Biol Chem 287(51):42995–43006. https://doi.org/10.1074/jbc.M112.392746
Damez-Werno DM, Sun H, Scobie KN, Shao N, Rabkin J, Dias C, Calipari ES, Maze I et al (2016) Histone arginine methylation in cocaine action in the nucleus accumbens. Proc Natl Acad Sci U S A 113(34):9623–9628. https://doi.org/10.1073/pnas.1605045113
Lee S-Y, Vuong TA, So H-K, Kim H-J, Kim YB, Kang J-S, Kwon I, Cho H (2020) PRMT7 deficiency causes dysregulation of the HCN channels in the CA1 pyramidal cells and impairment of social behaviors. Exp Mol Med 52(4):604–614. https://doi.org/10.1038/s12276-020-0417-x
Kousaka A, Mori Y, Koyama Y, Taneda T, Miyata S, Tohyama M (2009) The distribution and characterization of endogenous protein arginine N-methyltransferase 8 in mouse CNS. Neuroscience 163(4):1146–1157. https://doi.org/10.1016/j.neuroscience.2009.06.061
Taneda T, Miyata S, Kousaka A, Inoue K, Koyama Y, Mori Y, Tohyama M (2007) Specific regional distribution of protein arginine methyltransferase 8 (PRMT8) in the mouse brain. Brain Res 1155:1–9. https://doi.org/10.1016/j.brainres.2007.03.086
Dillon MB, Rust HL, Thompson PR, Mowen KA (2013) Automethylation of protein arginine methyltransferase 8 (PRMT8) regulates activity by impeding S-adenosylmethionine sensitivity. J Biol Chem 288(39):27872–27880. https://doi.org/10.1074/jbc.M113.491092
Mo C, Xu M, Wen C, Chang R, Huang C, Zou W, Zhu X, Guo Q (2018) Normalizing JMJD6 expression in rat spinal dorsal horn alleviates hyperalgesia following chronic constriction injury. Front Neurosci 12:542. https://doi.org/10.3389/fnins.2018.00542
Cimato TR, Tang J, Xu Y, Guarnaccia C, Herschman HR, Pongor S, Aletta JM (2002) Nerve growth factor-mediated increases in protein methylation occur predominantly at type I arginine methylation sites and involve protein arginine methyltransferase 1. J Neurosci Res 67(4):435–442. https://doi.org/10.1002/jnr.10123
Chittka A (2013) Differential regulation of SC1/PRDM4 and PRMT5 mediated protein arginine methylation by the nerve growth factor and the epidermal growth factor in PC12 cells. Neurosci Lett 550:87–92. https://doi.org/10.1016/j.neulet.2013.06.051
Ratovitski T, Arbez N, Stewart JC, Chighladze E, Ross CA (2015) PRMT5-mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington’s disease (HD). Cell Cycle 14(11):1716–1729. https://doi.org/10.1080/15384101.2015.1033595
Holmes B, Benavides-Serrato A, Saunders JT, Landon KA, Schreck AJ, Nishimura RN, Gera J (2019) The protein arginine methyltransferase PRMT5 confers therapeutic resistance to mTOR inhibition in glioblastoma. J Neurooncol 145(1):11–22. https://doi.org/10.1007/s11060-019-03274-0
Lu YF, Cai XL, Li ZZ, Lv J, Xiang YA, Chen JJ, Chen W-J, Sun W-Y et al (2018) LncRNA SNHG16 functions as an oncogene by sponging MiR-4518 and up-regulating PRMT5 expression in Glioma. Cell Physiol Biochem 45(5):1975–1985. https://doi.org/10.1159/000487974
Zheng D, Chen D, Lin F, Wang X, Lu L, Luo S, Chen J, Xu X (2020) LncRNA NNT-AS1 promote glioma cell proliferation and metastases through miR-494-3p/PRMT1 axis. Cell Cycle 19(13):1621–1631. https://doi.org/10.1080/15384101.2020.1762037
Gasperini L, Rossi A, Cornella N, Peroni D, Zuccotti P, Potrich V, Quattrone A, Macchi P (2018) The hnRNP RALY regulates PRMT1 expression and interacts with the ALS-linked protein FUS: implication for reciprocal cellular localization. Mol Biol Cell 29(26):3067–3081. https://doi.org/10.1091/mbc.E18-02-0108
Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR (1996) The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem 271(25):15034–15044. https://doi.org/10.1074/jbc.271.25.15034
Miyata S, Mori Y, Tohyama M (2008) PRMT1 and Btg2 regulates neurite outgrowth of Neuro2a cells. Neurosci Lett 445(2):162–165. https://doi.org/10.1016/j.neulet.2008.08.065
Sanchez G, Dury AY, Murray LM, Biondi O, Tadesse H, El Fatimy R, Kothary R, Charbonnier F et al (2013) A novel function for the survival motoneuron protein as a translational regulator. Hum Mol Genet 22(4):668–684. https://doi.org/10.1093/hmg/dds474
Hsieh MC, Ho YC, Lai CY, Wang HH, Yang PS, Cheng JK, Chen G-D, Ng S-C et al (2021) Blocking the spinal Fbxo3/CARM1/K(+) channel epigenetic silencing pathway as a strategy for neuropathic pain relief. Neurotherapeutics 18(2):1295–1315. https://doi.org/10.1007/s13311-020-00977-5
Feng Q, He B, Jung S-Y, Song Y, Qin J, Tsai SY, Tsai M-J, O’Malley BW (2009) Biochemical control of CARM1 enzymatic activity by phosphorylation. J Biol Chem 284(52):36167–36174. https://doi.org/10.1074/jbc.M109.065524
Lim CS, Alkon DL (2012) Protein kinase C stimulates HuD-mediated mRNA stability and protein expression of neurotrophic factors and enhances dendritic maturation of hippocampal neurons in culture. Hippocampus 22(12):2303–2319. https://doi.org/10.1002/hipo.22048
Huang T, Yang Y, Song X, Wan X, Wu B, Sastry N, Horbinski CM, Zeng C et al (2021) PRMT6 methylation of RCC1 regulates mitosis, tumorigenicity, and radiation response of glioblastoma stem cells. Mol Cell 81(6):1276-1291.e1279. https://doi.org/10.1016/j.molcel.2021.01.015
Wu CY, Couto ESA, Citadin CT, Clemons GA, Acosta CH, Knox BA, Grames MS, Rodgers KM et al (2021) Palmitic acid methyl ester inhibits cardiac arrest-induced neuroinflammation and mitochondrial dysfunction. Prostag Leukotr Ess 165:102227. https://doi.org/10.1016/j.plefa.2020.102227
Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE (2000) Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol 20(13):4859–4869. https://doi.org/10.1128/mcb.20.13.4859-4869.2000
Cimato TR, Ettinger MJ, Zhou X, Aletta JM (1997) Nerve growth factor-specific regulation of protein methylation during neuronal differentiation of PC12 cells. J Cell Biol 138(5):1089–1103. https://doi.org/10.1083/jcb.138.5.1089
Amano G, Matsuzaki S, Mori Y, Miyoshi K, Han S, Shikada S, Takamura H, Yoshimura T et al (2020) SCYL1 arginine methylation by PRMT1 is essential for neurite outgrowth via Golgi morphogenesis. Mol Biol Cell 31(18):1963–1973. https://doi.org/10.1091/mbc.E20-02-0100
Yoshimoto T, Boehm M, Olive M, Crook MF, San H, Langenickel T, Nabel EG (2006) The arginine methyltransferase PRMT2 binds RB and regulates E2F function. Exp Cell Res 312(11):2040–2053. https://doi.org/10.1016/j.yexcr.2006.03.001
Swiercz R, Cheng D, Kim D, Bedford MT (2007) Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J Biol Chem 282(23):16917–16923. https://doi.org/10.1074/jbc.M609778200
Bachand F, Silver PA (2004) PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. Life Sci Alliance 23(13):2641–2650. https://doi.org/10.1038/sj.emboj.7600265
Miyata S, Mori Y, Tohyama M (2010) PRMT3 is essential for dendritic spine maturation in rat hippocampal neurons. Brain Res 1352:11–20. https://doi.org/10.1016/j.brainres.2010.07.033
Selvi BR, Swaminathan A, Maheshwari U, Nagabhushana A, Mishra RK, Kundu TK (2015) CARM1 regulates astroglial lineage through transcriptional regulation of Nanog and posttranscriptional regulation by miR92a. Mol Biol Cell 26(2):316–326. https://doi.org/10.1091/mbc.E14-01-0019
O’Brien KB, Alberich-Jordà M, Yadav N, Kocher O, Diruscio A, Ebralidze A, Levantini E, Sng NJL et al (2010) CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development 137(13):2147–2156. https://doi.org/10.1242/dev.037150
Fujiwara T, Mori Y, Chu DL, Koyama Y, Miyata S, Tanaka H, Yachi K, Kubo T et al (2006) CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol Cell Biol 26(6):2273–2285. https://doi.org/10.1128/MCB.26.6.2273-2285.2006
Tee WW, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, Surani MA (2010) Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev 24(24):2772–2777. https://doi.org/10.1101/gad.606110
Sohail M, Zhang M, Litchfield D, Wang L, Kung S, Xie J (2015) Differential expression, distinct localization and opposite effect on Golgi structure and cell differentiation by a novel splice variant of human PRMT5. Biochim Biophys Acta 1853(10 Pt A):2444–2452. https://doi.org/10.1016/j.bbamcr.2015.07.003
Li X, Xiao J, Fröhlich H, Tu X, Li L, Xu Y, Cao H, Qu J et al (2015) Foxp1 regulates cortical radial migration and neuronal morphogenesis in developing cerebral cortex. PLoS One 10(5):e0127671. https://doi.org/10.1371/journal.pone.0127671
Chiang K, Zielinska AE, Shaaban AM, Sanchez-Bailon MP, Jarrold J, Clarke TL, Zhang J, Francis A et al (2017) PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep 21(12):3498–3513. https://doi.org/10.1016/j.celrep.2017.11.096
Chen Z, Gan J, Wei Z, Zhang M, Du Y, Xu C, Zhao H (2022) The emerging role of PRMT6 in cancer. Front Oncol 12:841381. https://doi.org/10.3389/fonc.2022.841381
Neault M, Mallette FA, Vogel G, Michaud-Levesque J, Richard S (2012) Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res 40(19):9513–9521. https://doi.org/10.1093/nar/gks764
Stein C, Nötzold RR, Riedl S, Bouchard C, Bauer UM (2016) The arginine methyltransferase PRMT6 cooperates with polycomb proteins in regulating HOXA gene expression. PLoS One 11(2):e0148892. https://doi.org/10.1371/journal.pone.0148892
Blanc RS, Vogel G, Chen T, Crist C, Richard S (2016) PRMT7 preserves satellite cell regenerative capacity. Cell Rep 14(6):1528–1539. https://doi.org/10.1016/j.celrep.2016.01.022
Dhar SS, Lee SH, Kan PY, Voigt P, Ma L, Shi X, Reinberg D, Lee MG (2012) Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev 26(24):2749–2762. https://doi.org/10.1101/gad.203356.112
Couto ESA, Wu CY, Clemons GA, Acosta CH, Chen CT, Possoit HE, Citadin CT, Lee RH et al (2021) Protein arginine methyltransferase 8 modulates mitochondrial bioenergetics and neuroinflammation after hypoxic stress. J Neurochem 159(4):742–761. https://doi.org/10.1111/jnc.15462
Kim JD, Park KE, Ishida J, Kako K, Hamada J, Kani S, Takeuchi M, Namiki K et al (2015) PRMT8 as a phospholipase regulates Purkinje cell dendritic arborization and motor coordination. Sci Adv 1(11):e1500615. https://doi.org/10.1126/sciadv.1500615
Lo LH, Dong R, Lyu Q, Lai KO (2020) The protein arginine methyltransferase PRMT8 and substrate G3BP1 control Rac1-PAK1 signaling and actin cytoskeleton for dendritic spine maturation. Cell Rep 31(10):107744. https://doi.org/10.1016/j.celrep.2020.107744
Lin YL, Tsai YJ, Liu YF, Cheng YC, Hung CM, Lee YJ, Pan H, Li C (2013) The critical role of protein arginine methyltransferase prmt8 in zebrafish embryonic and neural development is non-redundant with its paralogue prmt1. PLoS One 8(3):e55221. https://doi.org/10.1371/journal.pone.0055221
Simandi Z, Czipa E, Horvath A, Koszeghy A, Bordas C, Póliska S, Juhász I, Imre L et al (2015) PRMT1 and PRMT8 regulate retinoic acid-dependent neuronal differentiation with implications to neuropathology. Stem Cells 33(3):726–741. https://doi.org/10.1002/stem.1894
Hashimoto M, Hirata T, Yonekawa C, Takeichi K, Fukamizu A, Nakagawa T, Kizuka Y (2020) Region-specific upregulation of HNK-1 glycan in the PRMT1-deficient brain. Biochim Biophys Acta Gen Subj 1864(3):129509. https://doi.org/10.1016/j.bbagen.2019.129509
Kim JI, Ganesan S, Luo SX, Wu YW, Park E, Huang EJ, Chen L, Ding JB (2015) Aldehyde dehydrogenase 1a1 mediates a GABA synthesis pathway in midbrain dopaminergic neurons. Science 350(6256):102–106. https://doi.org/10.1126/science.aac4690
Verma M, Khan MIK, Kadumuri RV, Chakrapani B, Awasthi S, Mahesh A, Govindaraju G, Chavali PL et al (2021) PRMT3 interacts with ALDH1A1 and regulates gene-expression by inhibiting retinoic acid signaling. Commun Biol 4(1):109. https://doi.org/10.1038/s42003-020-01644-3
Likhite N, Jackson CA, Liang M-S, Krzyzanowski MC, Lei P, Wood JF, Birkaya B, Michaels KL et al (2015) The protein arginine methyltransferase PRMT5 promotes D2-like dopamine receptor signaling. Sci Signal 8(402):ra115. https://doi.org/10.1126/scisignal.aad0872
Bowitch A, Michaels KL, Yu MC, Ferkey DM (2018) The protein arginine methyltransferase PRMT-5 regulates SER-2 tyramine receptor-mediated behaviors in Caenorhabditis elegans. G3 (Bethesda) 8(7):2389–2398. https://doi.org/10.1534/g3.118.200360
Penney J, Seo J, Kritskiy O, Elmsaouri S, Gao F, Pao P-C, Su SC, Tsai L-H (2017) Loss of Protein arginine methyltransferase 8 alters synapse composition and function, resulting in behavioral defects. J Neurosci 37(36):8655–8666. https://doi.org/10.1523/jneurosci.0591-17.2017
Lee PK, Goh WW, Sng JC (2017) Network-based characterization of the synaptic proteome reveals that removal of epigenetic regulator Prmt8 restricts proteins associated with synaptic maturation. J Neurochem 140(4):613–628. https://doi.org/10.1111/jnc.13921
Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M et al (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell Proteomics 13(1):372–387. https://doi.org/10.1074/mcp.O113.027870
Lee S-Y, Vuong TA, Wen X, Jeong H-J, So H-K, Kwon I, Kang J-S, Cho H (2019) Methylation determines the extracellular calcium sensitivity of the leak channel NALCN in hippocampal dentate granule cells. Exp Mol Med 51(10):1–14. https://doi.org/10.1038/s12276-019-0325-0
Ma T, Li L, Chen R, Yang L, Sun H, Du S, Xu X, Cao Z et al (2022) Protein arginine methyltransferase 7 modulates neuronal excitability by interacting with NaV1.9. Pain 163(4):753–764. https://doi.org/10.1097/j.pain.0000000000002421
Baek J-H, Rubinstein M, Scheuer T, Trimmer JS (2014) Reciprocal changes in phosphorylation and methylation of mammalian brain sodium channels in response to Seizures. J Biol Chem 289(22):15363–15373. https://doi.org/10.1074/jbc.M114.562785
Ishino Y, Shimizu S, Tohyama M, Miyata S (2022) Coactivator-associated arginine methyltransferase 1 controls oligodendrocyte differentiation in the corpus callosum during early brain development. Dev Neurobiol 82(3):245–260. https://doi.org/10.1002/dneu.22871
Scaglione A, Patzig J, Liang J, Frawley R, Bok J, Mela A, Yattah C, Zhang J et al (2018) PRMT5-mediated regulation of developmental myelination. Nat Commun 9(1):2840. https://doi.org/10.1038/s41467-018-04863-9
Calabretta S, Vogel G, Yu Z, Choquet K, Darbelli L, Nicholson TB, Kleinman CL, Richard S (2018) Loss of PRMT5 promotes PDGFRα degradation during oligodendrocyte differentiation and myelination. Dev Cell 46(4):426-440.e425. https://doi.org/10.1016/j.devcel.2018.06.025
Dansu DK, Liang J, Selcen I, Zheng H, Moore DF, Casaccia P (2022) PRMT5 interacting partners and substrates in oligodendrocyte lineage cells. Front Cell Neurosci 16:820226. https://doi.org/10.3389/fncel.2022.820226
Ning H-m, Zhang Y-z, Han Y-y, Zhang C, Song X-p, Liang S, Yang X, Wang J (2022) Deletion of Prmt5 in cerebral endothelial cells leads to cerebrovascular disease and astrocyte activation. China Biotechnol 42(4):1–8. https://doi.org/10.13523/j.cb.2112026
Hashimoto M, Murata K, Ishida J, Kanou A, Kasuya Y, Fukamizu A (2016) Severe hypomyelination and developmental defects are caused in mice lacking protein arginine methyltransferase 1 (PRMT1) in the central nervous system. J Biol Chem 291(5):2237–2245. https://doi.org/10.1074/jbc.M115.684514
Hashimoto M, Kumabe A, Kim J-D, Murata K, Sekizar S, Williams A, Lu W, Ishida J et al (2021) Loss of PRMT1 in the central nervous system (CNS) induces reactive astrocytes and microglia during postnatal brain development. J Neurochem 156(6):834–847. https://doi.org/10.1111/jnc.15149
Honda M, Nakashima K, Katada S (2017) PRMT1 regulates astrocytic differentiation of embryonic neural stem/precursor cells. J Neurochem 142(6):901–907. https://doi.org/10.1111/jnc.14123
Chen L, Zhang M, Fang L, Yang X, Cao N, Xu L, Shi L, Cao Y (2021) Coordinated regulation of the ribosome and proteasome by PRMT1 in the maintenance of neural stemness in cancer cells and neural stem cells. J Biol Chem 297(5):101275. https://doi.org/10.1016/j.jbc.2021.101275
Hashimoto M, Takeichi K, Murata K, Kozakai A, Yagi A, Ishikawa K, Suzuki-Nakagawa C, Kasuya Y et al (2022) Regulation of neural stem cell proliferation and survival by protein arginine methyltransferase 1. Front Neurosci 16:948517. https://doi.org/10.3389/fnins.2022.948517
Chittka A (2010) Dynamic distribution of histone H4 arginine 3 methylation marks in the developing murine cortex. PLoS One 5(11):e13807. https://doi.org/10.1371/journal.pone.0013807
Zurita Rendón O, Silva Neiva L, Sasarman F, Shoubridge EA (2014) The arginine methyltransferase NDUFAF7 is essential for complex I assembly and early vertebrate embryogenesis. Hum Mol Genet 23(19):5159–5170. https://doi.org/10.1093/hmg/ddu239
Li MO, Sarkisian MR, Mehal WZ, Rakic P, Flavell RA (2003) Phosphatidylserine receptor is required for clearance of apoptotic cells. Science 302(5650):1560–1563. https://doi.org/10.1126/science.1087621
Zhang X, Gao Y, Lu L, Zhang Z, Gan S, Xu L, Lei A, Cao Y (2015) JmjC domain-containing protein 6 (Jmjd6) derepresses the transcriptional repressor transcription factor 7-like 1 (Tcf7l1) and is required for body axis patterning during Xenopus embryogenesis. J Biol Chem 290(33):20273–20283. https://doi.org/10.1074/jbc.M115.646554
Barak T, Ristori E, Ercan-Sencicek AG, Miyagishima DF, Nelson-Williams C, Dong W, Jin SC, Prendergast A et al (2021) PPIL4 is essential for brain angiogenesis and implicated in intracranial aneurysms in humans. Nat Med 27(12):2165–2175. https://doi.org/10.1038/s41591-021-01572-7
Yi J, Shen HF, Qiu JS, Huang MF, Zhang WJ, Ding JC, Zhu X-Y, Zhou Y et al (2017) JMJD6 and U2AF65 co-regulate alternative splicing in both JMJD6 enzymatic activity dependent and independent manner. Nucleic Acids Res 45(6):3503–3518. https://doi.org/10.1093/nar/gkw1144
Bai X, Sui C, Liu F, Chen T, Zhang L, Zheng Y, Liu B, Gao C (2022) The protein arginine methyltransferase PRMT9 attenuates MAVS activation through arginine methylation. Nat Commun 13(1):5016. https://doi.org/10.1038/s41467-022-32628-y
Harada N, Takagi T, Nakano Y, Yamaji R, Inui H (2015) Protein arginine methyltransferase 10 is required for androgen-dependent proliferation of LNCaP prostate cancer cells. Biosci Biotechnol Biochem 79(9):1430–1437. https://doi.org/10.1080/09168451.2015.1025035
Ju UI, Park JW, Park HS, Kim SJ, Chun YS (2015) FBXO11 represses cellular response to hypoxia by destabilizing hypoxia-inducible factor-1α mRNA. Biochem Biophys Res Commun 464(4):1008–1015. https://doi.org/10.1016/j.bbrc.2015.07.037
Wang S, Tan X, Yang B, Yin B, Yuan J, Qiang B, Peng X (2012) The role of protein arginine-methyltransferase 1 in gliomagenesis. BMB Rep 45(8):470–475. https://doi.org/10.5483/BMBRep.2012.45.8.022
Favia A, Salvatori L, Nanni S, Iwamoto-Stohl LK, Valente S, Mai A, Scagnoli F, Fontanella RA et al (2019) The Protein Arginine Methyltransferases 1 and 5 affect Myc properties in glioblastoma stem cells. Sci Rep 9(1):15925. https://doi.org/10.1038/s41598-019-52291-6
Hua ZY, Hansen JN, He M, Dai SK, Choi Y, Fulton MD, Lloyd SM, Szemes M et al (2020) PRMT1 promotes neuroblastoma cell survival through ATF5. Oncogenesis 9(5):50. https://doi.org/10.1038/s41389-020-0237-9
Liao Y, Luo Z, Lin Y, Chen H, Chen T, Xu L, Orgurek S, Berry K et al (2022) PRMT3 drives glioblastoma progression by enhancing HIF1A and glycolytic metabolism. Cell Death Dis 13(11):943. https://doi.org/10.1038/s41419-022-05389-1
Banasavadi-Siddegowda YK, Russell L, Frair E, Karkhanis VA, Relation T, Yoo JY, Zhang J, Sif S et al (2017) PRMT5-PTEN molecular pathway regulates senescence and self-renewal of primary glioblastoma neurosphere cells. Oncogene 36(2):263–274. https://doi.org/10.1038/onc.2016.199
Braun CJ, Stanciu M, Boutz PL, Patterson JC, Calligaris D, Higuchi F, Neupane R, Fenoglio S et al (2017) Coordinated splicing of regulatory detained introns within oncogenic transcripts creates an exploitable vulnerability in malignant Glioma. Cancer Cell 32(4):411-426.e411. https://doi.org/10.1016/j.ccell.2017.08.018
Du C, Hansen LJ, Singh SX, Wang F, Sun R, Moure CJ, Roso K, Greer PK et al (2019) A PRMT5-RNF168-SMURF2 axis controls H2AX proteostasis. Cell Rep 28(12):3199-3211.e3195. https://doi.org/10.1016/j.celrep.2019.08.031
Park JH, Szemes M, Vieira GC, Melegh Z, Malik S, Heesom KJ, Von Wallwitz-Freitas L, Greenhough A et al (2015) Protein arginine methyltransferase 5 is a key regulator of the MYCN oncoprotein in neuroblastoma cells. Mol Oncol 9(3):617–627. https://doi.org/10.1016/j.molonc.2014.10.015
Chaturvedi NK, Mahapatra S, Kesherwani V, Kling MJ, Shukla M, Ray S, Kanchan R, Perumal N et al (2019) Role of protein arginine methyltransferase 5 in group 3 (MYC-driven) Medulloblastoma. BMC Cancer 19(1):1056. https://doi.org/10.1186/s12885-019-6291-z
Feng J, Dang Y, Zhang W, Zhao X, Zhang C, Hou Z, Jin Y, McNutt MA et al (2019) PTEN arginine methylation by PRMT6 suppresses PI3K-AKT signaling and modulates pre-mRNA splicing. Proc Natl Acad Sci U S A 116(14):6868–6877. https://doi.org/10.1073/pnas.1811028116
Hernandez S, Dominko T (2016) Novel protein arginine methyltransferase 8 isoform is essential for cell proliferation. J Cell Biochem 117(9):2056–2066. https://doi.org/10.1002/jcb.25508
Miller TE, Liau BB, Wallace LC, Morton AR, Xie Q, Dixit D, Factor DC, Kim LJY et al (2017) Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature 547(7663):355–359. https://doi.org/10.1038/nature23000
Zhou DX, Zhou D, Zhan SQ, Wang P, Qin K, Gan W, Lin XF (2017) Inhibition of JMJD6 expression reduces the proliferation, migration and invasion of neuroglioma stem cells. Neoplasma 64(5):700–708. https://doi.org/10.4149/neo_2017_507
Clemons GA, Silva ACE, Acosta CH, Udo MSB, Tesic V, Rodgers KM, Wu CY-C, Citadin CT et al (2022) Protein arginine methyltransferase 4 modulates nitric oxide synthase uncoupling and cerebral blood flow in Alzheimer’s disease. J Cell Physiol. https://doi.org/10.1002/jcp.30858
Suganuma T, Swanson SK, Gogol M, Garrett TJ, Conkright-Fincham J, Florens L, Washburn MP, Workman JL (2018) MPTAC determines APP fragmentation via sensing sulfur amino acid catabolism. Cell Rep 24(6):1585–1596. https://doi.org/10.1016/j.celrep.2018.07.013
Ishii A, Matsuba Y, Mihira N, Kamano N, Saito T, Muramatsu SI, Yokosuka M, Saido TC et al (2022) Tau-binding protein PRMT8 facilitates vacuole degeneration in the brain. J Biochem 172(4):233–243. https://doi.org/10.1093/jb/mvac058
Migazzi A, Scaramuzzino C, Anderson EN, Tripathy D, Hernández IH, Grant RA, Roccuzzo M, Tosatto L et al (2021) Huntingtin-mediated axonal transport requires arginine methylation by PRMT6. Cell Rep 35(2):108980. https://doi.org/10.1016/j.celrep.2021.108980
Ratovitski T, Jiang M, O’Meally RN, Rauniyar P, Chighladze E, Faragó A, Kamath SV, Jin J et al (2022) Interaction of huntingtin with PRMTs and its subsequent arginine methylation affects HTT solubility, phase transition behavior and neuronal toxicity. Hum Mol Genet 31(10):1651–1672. https://doi.org/10.1093/hmg/ddab351
Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, Ruepp M-D, Simons M et al (2018) Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173(3):706-719.e713. https://doi.org/10.1016/j.cell.2018.03.004
Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A et al (2018) FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173(3):720-734.e715. https://doi.org/10.1016/j.cell.2018.03.056
Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B et al (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495(7442):467–473. https://doi.org/10.1038/nature11922
Ryan VH, Dignon GL, Zerze GH, Chabata CV, Silva R, Conicella AE, Amaya J, Burke KA et al (2018) Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol Cell 69(3):465-479.e467. https://doi.org/10.1016/j.molcel.2017.12.022
Chitiprolu M, Jagow C, Tremblay V, Bondy-Chorney E, Paris G, Savard A, Palidwor G, Barry FA et al (2018) A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat Commun 9(1):2794. https://doi.org/10.1038/s41467-018-05273-7
Kawahara D, Suzuki T, Nakaya T (2021) Cytoplasmic granule formation by FUS-R495X is attributable to arginine methylation in all Gly-rich, RGG1 and RGG2 domains. Genes Cells 26(3):190–197. https://doi.org/10.1111/gtc.12827
Simandi Z, Pajer K, Karolyi K, Sieler T, Jiang LL, Kolostyak Z, Sari Z, Fekecs Z et al (2018) Arginine methyltransferase PRMT8 provides cellular stress tolerance in aging motoneurons. J Neurosci 38(35):7683–7700. https://doi.org/10.1523/jneurosci.3389-17.2018
Hubers L, Valderrama-Carvajal H, Laframboise J, Timbers J, Sanchez G, Côté J (2011) HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum Mol Genet 20(3):553–579. https://doi.org/10.1093/hmg/ddq500
Tadesse H, Deschênes-Furry J, Boisvenue S, Côté J (2008) KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum Mol Genet 17(4):506–524. https://doi.org/10.1093/hmg/ddm327
Scaramuzzino C, Casci I, Parodi S, Lievens PMJ, Polanco MJ, Milioto C, Chivet M, Monaghan J et al (2015) Protein arginine methyltransferase 6 enhances polyglutamine-expanded androgen receptor function and toxicity in spinal and bulbar muscular atrophy. Neuron 85(1):88–100. https://doi.org/10.1016/j.neuron.2014.12.031
Nho JH, Park MJ, Park HJ, Lee JH, Choi JH, Oh SJ, Lee Y-J, Yu Y-B et al (2020) Protein arginine methyltransferase-1 stimulates dopaminergic neuronal cell death in a Parkinson’s disease model. Biochem Biophys Res Commun 530(2):389–395. https://doi.org/10.1016/j.bbrc.2020.05.016
Webb LM, Amici SA, Jablonski KA, Savardekar H, Panfil AR, Li L, Zhou W, Peine K et al (2017) PRMT5-selective inhibitors suppress inflammatory T cell responses and Experimental Autoimmune Encephalomyelitis. J Immunol 198(4):1439–1451. https://doi.org/10.4049/jimmunol.1601702
Li J, Zeng Q, Su W, Song M, Xie M, Mao L (2021) FBXO10 prevents chronic unpredictable stress-induced behavioral despair and cognitive impairment through promoting RAGE degradation. CNS Neurosci Ther 27(12):1504–1517. https://doi.org/10.1111/cns.13727
Kim HJ, Jeong MH, Kim KR, Jung CY, Lee SY, Kim H, Koh J, Vuong TA et al (2016) Protein arginine methylation facilitates KCNQ channel-PIP2 interaction leading to seizure suppression. Elife 5. https://doi.org/10.7554/eLife.17159
Li Y, Zhu R, Wang W, Fu D, Hou J, Ji S, Chen B, Hu Z et al (2015) Arginine methyltransferase 1 in the nucleus accumbens regulates behavioral effects of cocaine. J Neurosci 35(37):12890–12902. https://doi.org/10.1523/jneurosci.0246-15.2015
Mo K, Xu H, Gong H, Lei H, Wang Y, Guo W, Xu S, Tu W (2018) Dorsal root ganglia coactivator-associated arginine methyltransferase 1 contributes to peripheral nerve injury-induced pain hypersensitivities. Neuroscience 394:232–242. https://doi.org/10.1016/j.neuroscience.2018.10.038
Wen C, Xu M, Mo C, Cheng Z, Guo Q, Zhu X (2018) JMJD6 exerts function in neuropathic pain by regulating NF-κB following peripheral nerve injury in rats. Int J Mol Med 42(1):633–642. https://doi.org/10.3892/ijmm.2018.3613
Zheng K, Zhang Y, Zhang C, Ye W, Ye C, Tan X, Xiong Y (2022) PRMT8 attenuates cerebral Ischemia/reperfusion injury via modulating microglia activation and polarization to suppress neuroinflammation by upregulating Lin28a. ACS Chem Neurosci 13(7):1096–1104. https://doi.org/10.1021/acschemneuro.2c00096
Samuel SF, Marsden AJ, Deepak S, Rivero F, Greenman J, Beltran-Alvarez P (2018) Inhibiting arginine methylation as a tool to investigate cross-talk with methylation and acetylation post-translational modifications in a Glioblastoma cell Line. Proteomes 6(4). https://doi.org/10.3390/proteomes6040044
Wang S, Tan XC, Yang B, Yin B, Peng XZ (2012) Screening of substrates of protein arginine methyltransferase 1 in glioma. Chin Med Sci J 27(1):1–6. https://doi.org/10.1016/s1001-9294(12)60014-5
Han X, Li R, Zhang W, Yang X, Wheeler CG, Friedman GK, Province P, Ding Q et al (2014) Expression of PRMT5 correlates with malignant grade in gliomas and plays a pivotal role in tumor growth in vitro. J Neurooncol 118(1):61–72. https://doi.org/10.1007/s11060-014-1419-0
Mongiardi MP, Savino M, Bartoli L, Beji S, Nanni S, Scagnoli F, Falchetti ML, Favia A et al (2015) Myc and Omomyc functionally associate with the Protein arginine methyltransferase 5 (PRMT5) in glioblastoma cells. Sci Rep 5:15494. https://doi.org/10.1038/srep15494
Tan Z, Chen K, Wu W, Zhou Y, Zhu J, Wu G, Cao L, Zhang X et al (2018) Overexpression of HOXC10 promotes angiogenesis in human glioma via interaction with PRMT5 and upregulation of VEGFA expression. Theranostics 8(18):5143–5158. https://doi.org/10.7150/thno.27310
Hernandez SJ, Dolivo DM, Dominko T (2017) PRMT8 demonstrates variant-specific expression in cancer cells and correlates with patient survival in breast, ovarian and gastric cancer. Oncol Lett 13(3):1983–1989. https://doi.org/10.3892/ol.2017.5671
Rosager AM, Dahlrot RH, Sørensen MD, Bangsø JA, Hansen S, Kristensen BW (2021) The epigenetic regulator Jumonji domain-containing protein 6 (JMJD6) is highly expressed but not prognostic in IDH-wildtype Glioblastoma patients. J Neuropathol Exp Neurol 81(1):54–60. https://doi.org/10.1093/jnen/nlab124
Kater MSJ, Huffels CFM, Oshima T, Renckens NS, Middeldorp J, Boddeke EWGM, Smit AB, Eggen BJL et al (2022) Prevention of microgliosis halts early memory loss in a mouse model of Alzheimer’s disease. Brain Behav Immun. https://doi.org/10.1016/j.bbi.2022.10.009
Choi S, Singh I, Singh AK, Khan M, Won J (2020) Asymmetric dimethylarginine exacerbates cognitive dysfunction associated with cerebrovascular pathology. Faseb J 34(5):6808–6823. https://doi.org/10.1096/fj.201901318R
Cho EC, Zheng S, Munro S, Liu G, Carr SM, Moehlenbrink J, Lu Y-C, Stimson L et al (2012) Arginine methylation controls growth regulation by E2F–1. Embo J 31(7):1785–1797. https://doi.org/10.1038/emboj.2012.17
Rudich P, Watkins S, Lamitina T (2020) PolyQ-independent toxicity associated with novel translational products from CAG repeat expansions. PLoS One 15(4):e0227464. https://doi.org/10.1371/journal.pone.0227464
Scaramuzzino C, Monaghan J, Milioto C, Lanson NA Jr, Maltare A, Aggarwal T, Casci I, Fackelmayer FO et al (2013) Protein arginine methyltransferase 1 and 8 interact with FUS to modify its sub-cellular distribution and toxicity in vitro and in vivo. PLoS One 8(4):e61576. https://doi.org/10.1371/journal.pone.0061576
Zhou H, Mangelsdorf M, Liu J, Zhu L, Wu JY (2014) RNA-binding proteins in neurological diseases. Sci China Life Sci 57(4):432–444. https://doi.org/10.1007/s11427-014-4647-9
Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Velde CV, Bouchard J-P, Lacomblez L et al (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 40(5):572–574. https://doi.org/10.1038/ng.132
Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B et al (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323(5918):1208–1211. https://doi.org/10.1126/science.1165942
Ikenaka K, Atsuta N, Maeda Y, Hotta Y, Nakamura R, Kawai K, Yokoi D, Hirakawa A et al (2019) Increase of arginine dimethylation correlates with the progression and prognosis of ALS. Neurology 92(16):e1868–e1877. https://doi.org/10.1212/wnl.0000000000007311
Shaw PJ (2005) Molecular and cellular pathways of neurodegeneration in motor neurone disease. J Neurol Neurosurg Psychiatry 76(8):1046–1057. https://doi.org/10.1136/jnnp.2004.048652
Dong R, Li X, Lai KO (2021) Activity and function of the PRMT8 protein arginine methyltransferase in neurons. Life-Basel 11(11). https://doi.org/10.3390/life11111132
Zilio E, Piano V, Wirth B (2022) Mitochondrial dysfunction in Spinal Muscular Atrophy. Int J Mol Sci 23(18). https://doi.org/10.3390/ijms231810878
Giavazzi A, Setola V, Simonati A, Battaglia G (2006) Neuronal-specific roles of the Survival Motor Neuron protein: evidence from Survival Motor Neuron expression patterns in the developing human central nervous system. J Neuropathol Exp Neurol 65(3):267–277. https://doi.org/10.1097/01.jnen.0000205144.54457.a3
Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K et al (2016) SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 529(7584):48–53. https://doi.org/10.1038/nature16469
Grunseich C, Fischbeck KH (2020) Molecular pathogenesis of spinal bulbar muscular atrophy (Kennedy’s disease) and avenues for treatment. Curr Opin Neurol 33(5):629–634. https://doi.org/10.1097/wco.0000000000000856
Ogura Y, Sahashi K, Hirunagi T, Iida M, Miyata T, Katsuno M (2022) Mid1 is associated with androgen-dependent axonal vulnerability of motor neurons in spinal and bulbar muscular atrophy. Cell Death Dis 13(7):601. https://doi.org/10.1038/s41419-022-05001-6
Bientinesi R, Coluzzi S, Gavi F, Nociti V, Gandi C, Marino F, Moretto S, Mirabella M et al (2022) The impact of neurogenic lower urinary tract symptoms and erectile dysfunctions on marital relationship in men with Multiple Sclerosis: a single cohort study. J Clin Med 11(19). https://doi.org/10.3390/jcm11195639
Rund BR (2018) The research evidence for schizophrenia as a neurodevelopmental disorder. Scand J Psychol 59(1):49–58. https://doi.org/10.1111/sjop.12414
Fiorica PN, Wheeler HE (2019) Transcriptome association studies of neuropsychiatric traits in African Americans implicate PRMT7 in schizophrenia. PeerJ 7:e7778. https://doi.org/10.7717/peerj.7778
Tillotson R, Bird A (2020) The molecular basis of MeCP2 function in the brain. J Mol Biol 432(6):1602–1623. https://doi.org/10.1016/j.jmb.2019.10.004
Schmidt A, Frei J, Poetsch A, Chittka A, Zhang H, Aßmann C, Zhou W, Peine K et al (2022) MeCP2 heterochromatin organization is modulated by arginine methylation and serine phosphorylation. Front Cell Dev Biol 10:941493. https://doi.org/10.3389/fcell.2022.941493
Poquérusse J, Whitford W, Taylor J, Alburaiky S, Snell RG, Lehnert K, Jacobsen JC (2022) Novel PRMT7 mutation in a rare case of dysmorphism and intellectual disability. J Hum Genet 67(1):19–26. https://doi.org/10.1038/s10038-021-00955-5
Agolini E, Dentici ML, Bellacchio E, Alesi V, Radio FC, TorellaMusacchia AF, Tartaglia M et al (2018) Expanding the clinical and molecular spectrum of PRMT7 mutations: 3 additional patients and review. Clin Genet 93(3):675–681. https://doi.org/10.1111/cge.13137
Kernohan KD, McBride A, Xi Y, Martin N, Schwartzentruber J, Dyment DA, Majewski J, Blaser S et al (2017) Loss of the arginine methyltranserase PRMT7 causes syndromic intellectual disability with microcephaly and brachydactyly. Clin Genet 91(5):708–716. https://doi.org/10.1111/cge.12884
Birnbaum R, Yosha-Orpaz N, Yanoov-Sharav M, Kidron D, Gur H, Yosovich K, Lerman-Sagie T, Malinger G et al (2019) Prenatal and postnatal presentation of PRMT7 related syndrome: Expanding the phenotypic manifestations. Am J Med Genet A 179(1):78–84. https://doi.org/10.1002/ajmg.a.6
Franić S, Groen-Blokhuis MM, Dolan CV, Kattenberg MV, Pool R, Xiao X, Scheet PA, Ehli EA et al (2015) Intelligence: shared genetic basis between Mendelian disorders and a polygenic trait. Eur J Hum Genet 23(10):1378–1383. https://doi.org/10.1038/ejhg.2015.3
Hellmann-Regen J, Piber D, Hinkelmann K, Gold SM, Heesen C, Spitzer C, Endres M, Otte C (2013) Depressive syndromes in neurological disorders. Eur Arch Psychiatry Clin Neurosci 263(Suppl 2):S123-136. https://doi.org/10.1007/s00406-013-0448-6
Bozzi Y, Casarosa S, Caleo M (2012) Epilepsy as a neurodevelopmental disorder. Front Psychiatry 3:19. https://doi.org/10.3389/fpsyt.2012.00019
McCrory EJ, Mayes L (2015) Understanding Addiction as a developmental disorder: an argument for a developmentally informed multilevel approach. Curr Addict Rep 2(4):326–330. https://doi.org/10.1007/s40429-015-0079-2
Xu HL, Xu SY, Mo K (2017) Transcription of protein arginine N-methyltransferase genes in mouse dorsal root ganglia following peripheral nerve injury. Nan Fang Yi Ke Da Xue Xue Bao 37(12):1620–1625. https://doi.org/10.3969/j.issn.1673-4254.2017.12.10
Lin L, Wang X, Yu Z (2016) Ischemia-reperfusion Injury in the brain: mechanisms and potential therapeutic strategies. Biochem Pharmacol 5(4):213. https://doi.org/10.4172/2167-0501.1000213
Xu H, Zheng L, Chen X, Pang Q, Yan Y, Liu R, Guo H, Ren Z et al (2022) Brain-specific loss of Abcg1 disturbs cholesterol metabolism and aggravates pyroptosis and neurological deficits after traumatic brain injury. Brain Pathol 33:e13126. https://doi.org/10.1111/bpa.13126
Bao Z, Liu Y, Chen B, Miao Z, Tu Y, Li C, Chao H, Ye Y et al (2021) Prokineticin-2 prevents neuronal cell deaths in a model of traumatic brain injury. Nat Commun 12(1):4220. https://doi.org/10.1038/s41467-021-24469-y
Lochhead JJ, Ronaldson PT, Davis TP (2017) Hypoxic stress and inflammatory pain disrupt blood-brain barrier tight junctions: implications for drug delivery to the central nervous system. AAPS J 19(4):910–920. https://doi.org/10.1208/s12248-017-0076-6
Hamamoto R, Saloura V, Nakamura Y (2015) Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer 15(2):110–124. https://doi.org/10.1038/nrc3884
Chen Y, Zhang M, Wu A, Yao X, Wang Q (2022) Structure-based discovery and biological assays of a novel PRMT5 inhibitor for non-small cell lung cancer. Molecules 27(21). https://doi.org/10.3390/molecules27217436
Lee FYF, Wu W-L, Yang Z, Tan J (2021) Abstract 1145: AGX323 - A SAM-competitive, orally available inhibitor of protein arginine methyltransferase 5 (PRMT5) with potent cellular antiproliferative and in vivo antitumor activity against selected solid cancer types. Cancer Res 81(13_Supplement):1145–1145. https://doi.org/10.1158/1538-7445.Am2021-1145
McKinney DC, McMillan BJ, Ranaghan MJ, Moroco JA, Brousseau M, Mullin-Bernstein Z, O’Keefe M, McCarren P et al (2021) Discovery of a first-in-class inhibitor of the PRMT5-substrate adaptor interaction. J Med Chem 64(15):11148–11168. https://doi.org/10.1021/acs.jmedchem.1c00507
Ye Y, Zhang B, Mao R, Zhang C, Wang Y, Xing J, Liu Y-C, Luo X et al (2017) Discovery and optimization of selective inhibitors of protein arginine methyltransferase 5 by docking-based virtual screening. Org Biomol Chem 15(17):3648–3661. https://doi.org/10.1039/c7ob00070g
Zheng BN, Ding CH, Chen SJ, Zhu K, Shao J, Feng J, Xu W-P, Cai L-Y et al (2019) Targeting PRMT5 Activity Inhibits the Malignancy of Hepatocellular Carcinoma by Promoting the Transcription of HNF4α. Theranostics 9(9):2606–2617. https://doi.org/10.7150/thno.32344
Belmontes B, Policheni A, Liu S, Slemmons K, Moriguchi J, Ma H, Aiello D, Yang Y et al (2022) Abstract 1807: the discovery and preclinical characterization of the MTA cooperative PRMT5 inhibitor AM-9747. Cancer Res 82(12_Supplement):1807–1807. https://doi.org/10.1158/1538-7445.Am2022-1807
Gopalakrishnapillai A, Kisielewski A, Zhang Y, Ruggeri B, Scherle P, Kolb EA, Barwe SP (2021) Evaluating the efficacy of PRMT5 inhibitor C220 in patient-derived xenograft models of pediatric acute myeloid leukemia. Blood 138(Supplement 1):1170–1170. https://doi.org/10.1182/blood-2021-151671
Smil D, Eram MS, Li F, Kennedy S, Szewczyk MM, Brown PJ, Barsyte-Lovejoy D, Arrowsmith CH et al (2015) Discovery of a dual PRMT5-PRMT7 inhibitor. ACS Med Chem Lett 6(4):408–412. https://doi.org/10.1021/ml500467h
Gerhart SV, Kellner WA, Thompson C, Pappalardi MB, Zhang X-P, Montes de Oca R, Penebre E, Duncan K et al (2018) Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci Rep 8(1):9711. https://doi.org/10.1038/s41598-018-28002-y
Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, Rioux N, Munchhof MJ et al (2015) A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol 11(6):432–437. https://doi.org/10.1038/nchembio.1810
Bonday ZQ, Cortez GS, Grogan MJ, Antonysamy S, Weichert K, Bocchinfuso WP, Li F, Kennedy S et al (2018) LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med Chem Lett 9(7):612–617. https://doi.org/10.1021/acsmedchemlett.8b00014
Smith CR, Kulyk S, Lawson JD, Engstrom LD, Aranda R, Briere DM, Gunn R, Moya K et al (2021) Abstract LB003: Fragment based discovery of MRTX9768, a synthetic lethal-based inhibitor designed to bind the PRMT5-MTA complex and selectively target MTAP/CDKN2A-deleted tumors. Cancer Res 81(13_Supplement):LB003. https://doi.org/10.1158/1538-7445.Am2021-lb003
Shen Y, Gao G, Yu X, Kim H, Wang L, Xie L, Schwarz M, Chen X et al (2020) Discovery of first-in-class protein arginine methyltransferase 5 (PRMT5) degraders. J Med Chem 63(17):9977–9989. https://doi.org/10.1021/acs.jmedchem.0c01111
Mcalpine IJ, Tatlock J, Billitti J, Braganza J, Brooun A, Ya-Li D, Hirakawa B, Jensen-Pergakes K et al (2018) Abstract 4857: Discovery of PF-06855800, a SAM competitive PRMT5 inhibitor with potent antitumor activity. Cancer Res 78(13_Supplement):4857. https://doi.org/10.1158/1538-7445.Am2018-4857
Zhang P, Tao H, Yu L, Zhou L, Zhu C (2020) Developing protein arginine methyltransferase 1 (PRMT1) inhibitor TC-E-5003 as an antitumor drug using INEI drug delivery systems. Drug Deliv 27(1):491–501. https://doi.org/10.1080/10717544.2020.1745327
Tang S, Sethunath V, Metaferia NY, Nogueira MF, Gallant DS, Garner ER, Lairson LA, Penney CM et al (2022) A genome-scale CRISPR screen reveals PRMT1 as a critical regulator of androgen receptor signaling in prostate cancer. Cell Rep 38(8):110417. https://doi.org/10.1016/j.celrep.2022.110417
Kordala A, Ahlskog N, Hanifi M, Bhomra A, Stoodley J, Lim W, Hammond S, Wood M et al (2022) Type I PRMT inhibitor MS023 promotes SMN2 exon 7 inclusion and synergizes with nusinersen to rescue the phenotype of SMA mice. bioRxiv. https://doi.org/10.1101/2022.10.18.512489
Wang J, Wang C, Xu P, Li X, Lu Y, Jin D, Yin X, Jiang H et al (2021) PRMT1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Theranostics 11(11):5387–5403. https://doi.org/10.7150/thno.42345
Nakayama K, Szewczyk MM, Dela Sena C, Wu H, Dong A, Zeng H, Li F, de Freitas RF et al (2018) TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma. Oncotarget 9(26):18480–18493. https://doi.org/10.18632/oncotarget.24883
Drew AE, Moradei O, Jacques SL, Rioux N, Boriack-Sjodin AP, Allain C, Scott MP, Jin L et al (2017) Identification of a carm1 inhibitor with potent in vitro and in vivo activity in preclinical models of multiple myeloma. Sci Rep 7(1):17993. https://doi.org/10.1038/s41598-017-18446-z
Szewczyk MM, Ishikawa Y, Organ S, Sakai N, Li F, Halabelian L, Ackloo S, Couzens AL et al (2020) Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response. Nat Commun 11(1):2396. https://doi.org/10.1038/s41467-020-16271-z
Sachamitr P, Ho JC, Ciamponi FE, Ba-Alawi W, Coutinho FJ, Guilhamon P, Kushida MM, Cavalli FMG et al (2021) PRMT5 inhibition disrupts splicing and stemness in glioblastoma. Nat Commun 12(1):979. https://doi.org/10.1038/s41467-021-21204-5
Funding
This work was funded by the China Postdoctoral Science Foundation (2020M683510) and the Natural Science Foundation of Shaanxi Province, China (2022JQ-753).
Author information
Authors and Affiliations
Contributions
Shemin Lu had the idea for the article. Kewei Chang performed the literature search and wrote the manuscript. Kewei Chang, Liyan Lin and Tingting Cui prepared figures and tables. Shemin Lu, Dan Gao and Jidong Yan critically revised the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All authors give their consent for publication.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chang, K., Gao, D., Yan, J. et al. Critical Roles of Protein Arginine Methylation in the Central Nervous System. Mol Neurobiol 60, 6060–6091 (2023). https://doi.org/10.1007/s12035-023-03465-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-023-03465-x