Abstract
Background
As one of the most ubiquitous types of posttranscriptional modification, N6-methyladenosine (m6A) is extensively implicated in almost all types of cancers, including osteosarcoma. Our previous research partially uncovered the role of Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) in osteosarcoma. However, the relationships between methyltransferase-like 3 (METTL3) and noncoding RNAs modified by METTL3, especially MALAT1, in osteosarcoma remain obscure.
Methods
The expression of METTL3 in osteosarcoma was evaluated by online bioinformatics analysis, immunohistochemical (IHC) staining, western blotting (WB), and reverse transcription–quantitative PCR (RT‒qPCR). Cell Counting Kit 8 (CCK-8) and Transwell assays were used to evaluate the cell proliferation and invasion abilities. The expression of MALAT1 in osteosarcoma was evaluated by online bioinformatics analysis and RT‒qPCR analysis. m6A methylated RNA immunoprecipitation-qPCR (MeRIP-qPCR) was used to detect m6A modification changes in MALAT1. An actinomycin D assay was used to study changes in the stability of MALAT1.
Results
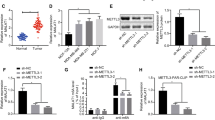
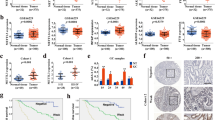
METTL3 was upregulated in osteosarcoma tissues and cell lines. Functionally, METTL3 promoted the proliferation and migration of osteosarcoma cells. Moreover, a clear positive correlation was found between METTL3 and MALAT1 expression, and MALAT1 was upregulated in osteosarcoma tissues and cells. Mechanistically, the presence of m6A modification sites in MALAT1 and METTL3-mediated m6A modification increased the stability of MALAT1 in osteosarcoma cells and promoted their proliferation and migration.
Conclusion
In this study, it was concluded that in osteosarcoma cells, METTL3, acting as an oncogene, promoted m6A modification of MALAT1, increased the stability of MALAT, and enhanced MALAT1-mediated oncogenic function.




Similar content being viewed by others
Data Availability
All the data involved in this study are listed.
References
Moore, D. D., & Luu, H. H. (2014). Osteosarcoma Cancer Treat Res, 162: 65–92.
Simpson, E., & Brown, H. L. (2018). Understanding osteosarcomas. Jaapa, 31(8), 15–19.
Meng, Y., et al. (2021). Circular RNA circNRIP1 plays oncogenic roles in the progression of osteosarcoma. Mammalian Genome, 32(6), 448–456.
Ianniello, Z., Paiardini, A., & Fatica, A. (2019). N(6)-Methyladenosine (m(6)A): A Promising New Molecular Target in Acute Myeloid Leukemia. Frontiers in Oncology, 9, 251.
Karthiya, R., & Khandelia, P. (2020). m6A RNA methylation: Ramifications for gene expression and Human Health. Molecular Biotechnology, 62(10), 467–484.
Meyer, K. D., et al. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell, 149(7), 1635–1646.
Bokar, J. A., et al. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. Rna, 3(11), 1233–1247.
Zeng, C., & Huang, W. (2020). Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. 13(1): p. 117.
Zhang, W., et al. (2020). Multifaceted functions and Novel Insight into the Regulatory Role of RNA N(6)-Methyladenosine modification in Musculoskeletal disorders. Front Cell Dev Biol, 8, 870.
Ling, Z., Chen, L., & Zhao, J. (2020). m6A-dependent up-regulation of DRG1 by METTL3 and ELAVL1 promotes growth, migration, and colony formation in osteosarcoma Biosci Rep, 40(4).
Wang, J., et al. (2022). m6A-dependent upregulation of TRAF6 by METTL3 is associated with metastatic osteosarcoma. J Bone Oncol, 32, 100411.
Wei, K., et al. (2022). Methylation recognition protein YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) regulates the proliferation, migration and invasion of osteosarcoma by regulating m6A level of CCR4-NOT transcription complex subunit 7 (CNOT7). Bioengineered, 13(3), 5236–5250.
Zhou, C., et al. (2020). N6-Methyladenosine modification of the TRIM7 positively regulates tumorigenesis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. EBioMedicine, 59, 102955.
Zhou, X., et al. (2021). METTL3 contributes to Osteosarcoma Progression by increasing DANCR mRNA Stability via m6A modification. Front Cell Dev Biol, 9, 784719.
Farzaneh, M., et al. (2023). LncRNA MALAT1-related signaling pathways in osteosarcoma. Clinical and Translational Oncology, 25(1), 21–32.
Malakoti, F., et al. (2021). Multiple function of lncRNA MALAT1 in cancer occurrence and progression. Chem Biol Drug Des.
Wang, Y., et al. (2017). Long non-coding RNA MALAT1 for promoting Metastasis and proliferation by acting as a ceRNA of mir-144-3p in osteosarcoma cells. Oncotarget, 8(35), 59417–59434.
Tao, L., et al. (2021). FTO modifies the m6A level of MALAT and promotes Bladder cancer progression. Oral Diseases, 11(2), e310.
Li, J., Momen-Heravi, F., & Wu, X. (2023). Mechanism of METTL14 and m6A modification of lncRNA MALAT1 in the proliferation of oral squamous cell carcinoma cells. Clin Transl Med, 29(5), 2012–2026.
Jiang, R., et al. (2022). METTL3 stabilizes HDAC5 mRNA in an m(6)A-dependent manner to facilitate malignant proliferation of osteosarcoma cells. Cell Death Discov, 8(1), 179.
Zeng, K., et al. (2018). CircHIPK3 promotes Colorectal cancer growth and Metastasis by sponging miR-7. Cell Death and Disease, 9(4), 417.
Chen, S., et al. (2019). TCM therapies combined with chemotherapy for preventing recurrence and Metastasis in postoperative II to IIIA NSCLC: A protocol for a systematic review and meta-analysis. Medicine (Baltimore), 98(9), e14724.
Wang, Y., et al. (2018). Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and Metastasis via decoying of mir-335-5p and miR-1972 in osteosarcoma. Molecular Cancer, 17(1), 89.
Wang, Y., et al. (2017). Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of mir-335-5p in osteosarcoma cells. Cancer Science, 108(5), 859–867.
Zhou, Y., et al. (2023). Silencing of IRF8 mediated by m6A modification promotes the progression of T-Cell Acute Lymphoblastic Leukemia. Adv Sci (Weinh), 10(2), e2201724.
Ratnadiwakara, M., & Änkö, M. L. (2018). mRNA Stability Assay using transcription inhibition by actinomycin D in mouse pluripotent stem cells. Bio Protoc, 8(21), e3072.
Barrett, T., et al. (2013). NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Research, 41(Database issue), D991–D995.
Chandrashekar, D. S., et al. (2022). UALCAN: An update to the integrated cancer data analysis platform. Neoplasia (New York, N.Y.), 25, 18–27.
Chandrashekar, D. S., et al. (2017). UALCAN: A portal for facilitating Tumor Subgroup Gene expression and survival analyses. Neoplasia (New York, N.Y.), 19(8), 649–658.
Tang, Z., et al. (2019). GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Research, 47(W1), W556–w560.
Zhou, Y., et al. (2016). SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Research, 44(10), e91.
Xuan, J. J., et al. (2018). RMBase v2.0: Deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Research, 46(D1), D327–d334.
Roundtree, I. A., et al. (2017). Dynamic RNA modifications in Gene expression regulation. Cell, 169(7), 1187–1200.
Liu, N., et al. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature, 518(7540), 560–564.
Wang, X., et al. (2015). N(6)-methyladenosine modulates Messenger RNA translation efficiency. Cell, 161(6), 1388–1399.
Wang, X., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature, 505(7481), 117–120.
Alarcón, C. R., et al. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature, 519(7544), 482–485.
Deng, X., et al. (2018). RNA N(6)-methyladenosine modification in cancers: Current status and perspectives. Cell Research, 28(5), 507–517.
Zhao, W., et al. (2020). Epigenetic regulation of m(6)a modifications in Human Cancer. Mol Ther Nucleic Acids, 19, 405–412.
Zeng, C., et al. (2020). Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. Journal of Hematology & Oncology, 13(1), 117.
Wu, Y., et al. (2022). The role of m6A methylation in osteosarcoma biological processes and its potential clinical value. Human Genomics, 16(1), 12.
Han, Y. (2019). CVm6A: A visualization and exploration database for m6As in cell lines. Cells, 8(2).
Zhang, Z., et al. (2021). N6-Methyladenosine-sculpted Regulatory Landscape of noncoding RNA. Frontiers in Oncology, 11, 743990.
Chen, L., et al. (2022). METTL3-mediated m6A modification stabilizes TERRA and maintains telomere stability. Nucleic Acids Research, 50(20), 11619–11634.
Ni, W., et al. (2019). Long noncoding RNA GAS5 inhibits progression of Colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)a reader YTHDF3. Molecular Cancer, 18(1), 143.
Zhang, H., et al. (2022). m6A methyltransferase METTL3-induced lncRNA SNHG17 promotes lung adenocarcinoma gefitinib resistance by epigenetically repressing LATS2 expression. Cell Death and Disease, 13(7), 657.
Acknowledgements
None.
Funding
None.
Author information
Authors and Affiliations
Contributions
YZ, YX and GQ were responsible for the performance of the experiments; YZ was responsible for the manuscript writing, figure completion and revision of the research. YL, YB, JL and TW were responsible for statistical analysis of the data. YW was mainly responsible for the scientific research and study design.
Corresponding authors
Ethics declarations
Patient Consent for Publication
Not applicable.
Ethical Approval and Consent to Participate
The Medical Ethics Committee of Central Hospital Affiliated with Shenyang Medical College approved this study (approval number: 2019DEC12-4), and all patients provided written informed consent.
Competing Interests
All authors state the absence of competing interests in the research and writing of the article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Xu, Y., Qiu, G. et al. METTL3 Mediated MALAT1 m6A Modification Promotes Proliferation and Metastasis in Osteosarcoma Cells. Mol Biotechnol (2023). https://doi.org/10.1007/s12033-023-00953-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-023-00953-2