Abstract
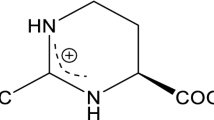
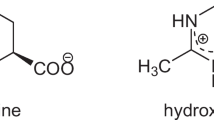
Ectoine and its derivative 5-hydroxyectoine are compatible solutes initially found in the hyperhalophilic bacterium Ectothiorhodospira halochloris, which inhabits the desert in Egypt. The habitat of ectoine producers implies the primary function of ectoine as a cytoprotectant against harsh conditions such as high salinity, drought, and high radiation. More extensive and in-depth studies have revealed the multiple functions of ectoine in its native producer bacterial cells and other types of cells and its biomolecular components (such as proteins and DNA) as a general protective agent. Its chemical properties as a bio-based amino acid derivative make it attractive for basic scientific research and related industries, such as the food/agricultural industry, cosmetic manufacturing, biologics, and therapeutic agent preparation. This article first discusses the functions and applications of ectoine and 5-hydroxyectoine. Subsequently, more emphasis was placed on advances in bio-based ectoine and/or 5-hydroxyectoine production. Strategies for developing more robust cell factories for highly efficient ectoine and/or 5-hydroxyectoine production are further discussed. We hope this review will provide a valuable reference for studies on the bio-based production of ectoine and 5-hydroxyectoine.




Similar content being viewed by others
References
Alsharif, W., Saad, M. M., & Hirt, H. (2020). Desert microbes for boosting sustainable agriculture in extreme environments. Frontiers in Microbiology, 11, 1666.
Somayaji, A., Dhanjal, C. R., Lingamsetty, R., Vinayagam, R., Selvaraj, R., Varadavenkatesan, T., & Govarthanan, M. (2022). An insight into the mechanisms of homeostasis in extremophiles. Microbiological Research, 263, 127115.
Afifa, H. N., Baqar, Z., Mumtaz, M., El-Sappah, A. H., Show, P. L., Iqbal, H. M. N., Varjani, S., & Bilal, M. (2022). Bioprospecting fungal-derived value-added bioproducts for sustainable pharmaceutical applications. Sustainable Chemistry and Pharmacy, 29, 100755.
Empadinhas, N., & da Costa, M. S. (2006). Diversity and biosynthesis of compatible solutes in hyper/thermophiles. International Microbiology, 9, 199–206.
Peters, P. (1990). The biosynthesis of ectoine. FEMS Microbiology Letters, 71, 157–162.
Graf, R., Anzali, S., Buenger, J., Pfluecker, F., & Driller, H. (2008). The multifunctional role of ectoine as a natural cell protectant. Clinics in Dermatology, 26, 326–333.
Ng, H. S., Wan, P.-K., Kondo, A., Chang, J.-S., & Lan, J.C.-W. (2023). Production and recovery of ectoine: A review of current state and future prospects. Processes, 11, 339.
Alves, A., Sousa, E., Kijjoa, A., & Pinto, M. (2020). Marine-derived compounds with potential use as cosmeceuticals and nutricosmetics. Molecules, 25, 2536.
Klein, J., Schwarz, T., & Lentzen, G. (2007). Ectoine as a natural component of food: Detection in red smear cheeses. Journal of Dairy Research, 74, 446–451.
Nakayama, H., Yoshida, K., Ono, H., Murooka, Y., & Shinmyo, A. (2000). Ectoine, the compatible solute of Halomonas elongata, confers hyperosmotic tolerance in cultured tobacco cells. Plant Physiology, 122, 1239–1247.
Boroujeni, M. B., & Nayeri, H. (2018). Stabilization of bovine lactoperoxidase in the presence of ectoine. Food Chemistry, 265, 208–215.
Kauth, M., & Trusova, O. V. (2022). Topical ectoine application in children and adults to treat inflammatory diseases associated with an impaired skin barrier: A systematic review. Dermatology and Therapy, 12, 295–313.
Becker, J., & Wittmann, C. (2020). Microbial production of extremolytes—High-value active ingredients for nutrition, health care, and well-being. Current Opinion in Biotechnology, 65, 118–128.
Held, C., Neuhaus, T., & Sadowski, G. (2010). Compatible solutes: Thermodynamic properties and biological impact of ectoines and prolines. Biophysical Chemistry, 152, 28–39.
Czech, L., Hermann, L., Stoveken, N., Richter, A. A., Hoppner, A., Smits, S. H. J., Heider, J., & Bremer, E. (2018). Role of the extremolytes ectoine and hydroxyectoine as stress protectants and nutrients: Genetics, phylogenomics, biochemistry, and structural analysis. Genes (Basel), 9(4), 177.
Jebbar, M., Talibart, R., Gloux, K., Bernard, T., & Blanco, C. (1992). Osmoprotection of Escherichia coli by ectoine: Uptake and accumulation characteristics. Journal of Bacteriology, 174, 5027–5035.
Rai, M., Pal, M., Sumesh, K. V., Jain, V., & Sankaranarayanan, A. (2006). Engineering for biosynthesis of ectoine (2-methyl 4-carboxy tetrahydro pyrimidine) in tobacco chloroplasts leads to accumulation of ectoine and enhanced salinity tolerance. Plant Science, 170, 291–306.
Bownik, A., & Stepniewska, Z. (2016). Ectoine as a promising protective agent in humans and animals. Archives of Industrial Hygiene and Toxicology, 67, 260–265.
Qiu, X., Yu, L., Cao, X., Wu, H., Xu, G., & Tang, X. (2021). Halomonas sedimenti sp. nov., a halotolerant bacterium isolated from deep-sea sediment of the southwest Indian Ocean. Current Microbiology, 78, 1662–1669.
Killian, M. S., Taylor, A. J., & Castner, D. G. (2018). Stabilization of dry protein coatings with compatible solutes. Biointerphases, 13, 06E401.
Van-Thuoc, D., Hashim, S. O., Hatti-Kaul, R., & Mamo, G. (2013). Ectoine-mediated protection of enzyme from the effect of pH and temperature stress: A study using Bacillus halodurans xylanase as a model. Applied Microbiology and Biotechnology, 97, 6271–6278.
Hahn, M. B., Meyer, S., Schroter, M. A., Kunte, H. J., Solomun, T., & Sturm, H. (2017). DNA protection by ectoine from ionizing radiation: Molecular mechanisms. Physical Chemistry Chemical Physics, 19, 25717–25722.
Schroter, M. A., Meyer, S., Hahn, M. B., Solomun, T., Sturm, H., & Kunte, H. J. (2017). Ectoine protects DNA from damage by ionizing radiation. Scientific Reports, 7, 15272.
Jorge, C. D., Borges, N., Bagyan, I., Bilstein, A., & Santos, H. (2016). Potential applications of stress solutes from extremophiles in protein folding diseases and healthcare. Extremophiles, 20, 251–259.
Kanapathipillai, M., Lentzen, G., Sierks, M., & Park, C. B. (2005). Ectoine and hydroxyectoine inhibit aggregation and neurotoxicity of Alzheimer’s beta-amyloid. FEBS Letters, 579, 4775–4780.
Sydlik, U., Gallitz, I., Albrecht, C., Abel, J., Krutmann, J., & Unfried, K. (2009). The compatible solute ectoine protects against nanoparticle-induced neutrophilic lung inflammation. American Journal of Respiratory and Critical Care Medicine, 180, 29–35.
Talibart, R., Jebbar, M., Gouesbet, G., Himdi-Kabbab, S., Wroblewski, H., Blanco, C., & Bernard, T. (1994). Osmoadaptation in rhizobia: Ectoine-induced salt tolerance. Journal of Bacteriology, 176, 5210–5217.
Tanne, C., Golovina, E. A., Hoekstra, F. A., Meffert, A., & Galinski, E. A. (2014). Glass-forming property of hydroxyectoine is the cause of its superior function as a desiccation protectant. Frontiers in Microbiology, 5, 150.
Manzanera, M., Garcia de Castro, A., Tondervik, A., Rayner-Brandes, M., Strom, A. R., & Tunnacliffe, A. (2002). Hydroxyectoine is superior to trehalose for anhydrobiotic engineering of Pseudomonas putida KT2440. Applied Environmental Microbiology, 68, 4328–4333.
Manzanera, M., Vilchez, S., & Tunnacliffe, A. (2004). High survival and stability rates of Escherichia coli dried in hydroxyectoine. FEMS Microbiology Letters, 233, 347–352.
Elbialy, H., Omara, A., Sharaf, A. M., El-Hela, A., Shahin, A., & El-Fouly, M. (2019). Isolation and characterization of new ectoine-producers from various hypersaline ecosystems in Egypt. Journal of Nuclear Technology in Applied Science, 7, 221–236.
Hermann, L., Mais, C. N., Czech, L., Smits, S. H. J., Bange, G., & Bremer, E. (2020). The ups and downs of ectoine: Structural enzymology of a major microbial stress protectant and versatile nutrient. Biological Chemistry, 401, 1443–1468.
Widderich, N., Hoppner, A., Pittelkow, M., Heider, J., Smits, S. H., & Bremer, E. (2014). Biochemical properties of ectoine hydroxylases from extremophiles and their wider taxonomic distribution among microorganisms. PLoS ONE, 9, e93809.
Wohlfarth, A., Severin, J., & Galinski, E. A. (1990). The spectrum of compatible solutes in heterotrophic halophilic eubacteria of the family Halomonadaceae. Journal of General Microbiology, 136, 705–712.
Vasistha, S., Khanra, A., Rai, M. P., Khan, S. A., Ma, Z., Munawaroh, H. S. H., Tang, D. Y. Y., & Show, P. L. (2023). Exploring the pivotal significance of microalgae-derived sustainable lipid production: A critical review of green bioenergy development. Energies, 16, 531.
Tang, D. Y. Y., Chu, D.-T., Nhi-Cong, L. T., Ratchahat, S., Banat, F., Chew, K. W., Gentili, F. G., & Show, P. L. (2023). Investigation and screening of mixed microalgae species for lipase production and recovery using liquid biphasic flotation approach. Journal of the Taiwan Institute of Chemical Engineers, 142, 104646.
Watthanasakphuban, N., Nguyen, L. V., Cheng, Y.-S., Show, P.-L., Sriariyanun, M., Koffas, M., & Rattanaporn, K. (2023). Development of a molasses-based medium for Agrobacterium tumefaciens fermentation for application in plant-based recombinant protein production. Fermentation, 9, 149.
Kunte, H., Lentzen, G., & Galinski, E. (2014). Industrial production of the cell protectant ectoine: Protection mechanisms, processes, and products. Current Biotechnology, 3, 10–25.
Onraedt, A. E., Walcarius, B. A., Soetaert, W. K., & Vandamme, E. J. (2005). Optimization of ectoine synthesis through fed-batch fermentation of Brevibacterium epidermis. Biotechnology Progress, 21, 1206–1212.
Zhang, L. H., Lang, Y. J., & Nagata, S. (2009). Efficient production of ectoine using ectoine-excreting strain. Extremophiles, 13, 717–724.
Chen, W. C., Hsu, C. C., Lan, J. C., Chang, Y. K., Wang, L. F., & Wei, Y. H. (2018). Production and characterization of ectoine using a moderately halophilic strain Halomonas salina BCRC17875. Journal of Bioscience and Bioengineering, 125, 578–584.
Chen, W. C., Hsu, C. C., Wang, L. F., Lan, J. C., Chang, Y. K., & Wei, Y. H. (2019). Exploring useful fermentation strategies for the production of hydroxyectoine with a halophilic strain, Halomonas salina BCRC 17875. Journal of Bioscience and Bioengineering, 128, 332–336.
Zhao, Q., Meng, Y., Li, S., Lv, P., Xu, P., & Yang, C. (2018). Genome sequence of Halomonas hydrothermalis Y2, an efficient ectoine-producer isolated from pulp mill wastewater. Journal of Biotechnology, 285, 38–41.
Zhao, Q., Li, S., Lv, P., Sun, S., Ma, C., Xu, P., Su, H., & Yang, C. (2019). High ectoine production by an engineered Halomonas hydrothermalis Y2 in a reduced salinity medium. Microbial Cell Factories, 18, 184.
Liu, M., Liu, H., Shi, M., Jiang, M., Li, L., & Zheng, Y. (2021). Microbial production of ectoine and hydroxyectoine as high-value chemicals. Microbial Cell Factories, 20, 76.
Chen, G. Q., Zhang, X., Liu, X., Huang, W., Xie, Z., Han, J., Xu, T., Mitra, R., Zhou, C., Zhang, J., & Chen, T. (2022). Halomonas spp., as chassis for low-cost production of chemicals. Applied Microbiology and Biotechnology, 106, 6977–6992.
Weinstock, M. T., Hesek, E. D., Wilson, C. M., & Gibson, D. G. (2016). Vibrio natriegens as a fast-growing host for molecular biology. Nature Methods, 13, 849–851.
Yin, J., Chen, J. C., Wu, Q., & Chen, G. Q. (2015). Halophiles, coming stars for industrial biotechnology. Biotechnology Advances, 33, 1433–1442.
Tan, S. I., Hsiang, C. C., & Ng, I. S. (2021). Tailoring genetic elements of the plasmid-driven T7 system for stable and robust one-step cloning and protein expression in broad Escherichia coli. ACS Synthetic Biology, 10, 2753–2762.
Kushwaha, M., & Salis, H. M. (2015). A portable expression resource for engineering cross-species genetic circuits and pathways. Nature Communications, 6, 7832.
Zhang, H., Liang, Z., Zhao, M., Ma, Y., Luo, Z., Li, S., & Xu, H. (2022). Metabolic engineering of Escherichia coli for ectoine production with a fermentation strategy of supplementing the amino donor. Frontiers in Bioengineering and Biotechnology, 10, 824859.
Hillier, H. T., Altermark, B., & Leiros, I. (2020). The crystal structure of the tetrameric DABA-aminotransferase EctB, a rate-limiting enzyme in the ectoine biosynthesis pathway. The FEBS Journal, 287, 4641–4658.
Ma, Q., Xia, L., Wu, H., Zhuo, M., Yang, M., Zhang, Y., Tan, M., Zhao, K., Sun, Q., Xu, Q., Chen, N., & Xie, X. (2022). Metabolic engineering of Escherichia coli for efficient osmotic stress-free production of compatible solute hydroxyectoine. Biotechnology and Bioengineering, 119, 89–101.
Kumar, D., & Gomes, J. (2005). Methionine production by fermentation. Biotechnology Advances, 23, 41–61.
Jiang, A., Song, Y., You, J., Zhang, X., Xu, M., & Rao, Z. (2022). High-yield ectoine production in engineered Corynebacterium glutamicum by fine metabolic regulation via plug-in repressor library. Bioresource Technology, 362, 127802.
Becker, J., Klopprogge, C., Herold, A., Zelder, O., Bolten, C. J., & Wittmann, C. (2007). Metabolic flux engineering of l-lysine production in Corynebacterium glutamicum—Over expression and modification of G6P dehydrogenase. Journal of Biotechnology, 132, 99–109.
Becker, J., Schafer, R., Kohlstedt, M., Harder, B. J., Borchert, N. S., Stoveken, N., Bremer, E., & Wittmann, C. (2013). Systems metabolic engineering of Corynebacterium glutamicum for production of the chemical chaperone ectoine. Microbial Cell Factories, 12, 110.
Kalinowski, J., Bathe, B., Bartels, D., Bischoff, N., Bott, M., Burkovski, A., Dusch, N., Eggeling, L., Eikmanns, B. J., Gaigalat, L., Goesmann, A., Hartmann, M., Huthmacher, K., Kramer, R., Linke, B., McHardy, A. C., Meyer, F., Mockel, B., Pfefferle, W., … Tauch, A. (2003). The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. Journal of Biotechnology, 104, 5–25.
Manfrao-Netto, J. H. C., Gomes, A. M. V., & Parachin, N. S. (2019). Advances in using Hansenula polymorpha as chassis for recombinant protein production. Frontiers in Bioengineering and Biotechnology, 7, 94.
Eilert, E., Kranz, A., Hollenberg, C. P., Piontek, M., & Suckow, M. (2013). Synthesis and release of the bacterial compatible solute 5-hydroxyectoine in Hansenula polymorpha. Journal of Biotechnology, 167, 85–93.
He, Y. Z., Gong, J., Yu, H. Y., Tao, Y., Zhang, S., & Dong, Z. Y. (2015). High production of ectoine from aspartate and glycerol by use of whole-cell biocatalysis in recombinant Escherichia coli. Microbial Cell Factories, 14, 55.
Wang, T., Ma, X., Du, G., & Chen, J. (2012). Overview of regulatory strategies and molecular elements in metabolic engineering of bacteria. Molecular Biotechnology, 52, 300–308.
Zhao, J., Qin, B., Nikolay, R., Spahn, C. M. T., & Zhang, G. (2019). Translatomics: The global view of translation. International Journal of Molecular Sciences, 20(1), 212.
Zhu, T., Guo, M., Zhuang, Y., Chu, J., & Zhang, S. (2011). Understanding the effect of foreign gene dosage on the physiology of Pichia pastoris by transcriptional analysis of key genes. Applied Microbiology and Biotechnology, 89, 1127–1135.
Shao, Z., Deng, W., Li, S., He, J., Ren, S., Huang, W., Lu, Y., Zhao, G., Cai, Z., & Wang, J. (2015). GlnR-mediated regulation of ectABCD transcription expands the role of the GlnR regulon to osmotic stress management. Journal of Bacteriology, 197, 3041–3047.
Zhang, D., Guan, D., Liang, J., Guo, C., Xie, X., Zhang, C., Xu, Q., & Chen, N. (2014). Reducing lactate secretion by ldhA Deletion in l-glutamate-producing strain Corynebacterium glutamicum GDK-9. Brazilian Journal of Microbiology, 45, 1477–1483.
Liu, Y., Xu, Y., Ding, D., Wen, J., Zhu, B., & Zhang, D. (2018). Genetic engineering of Escherichia coli to improve l-phenylalanine production. BMC Biotechnology, 18, 5.
Wang, Z., Qin, Q., Zheng, Y., Li, F., Zhao, Y., & Chen, G. Q. (2021). Engineering the permeability of Halomonas bluephagenesis enhanced its chassis properties. Metabolic Engineering, 67, 53–66.
Li, R., Wang, M., Ren, Z., Ji, Y., Yin, M., Zhou, H., & Tang, S. K. (2021). Amycolatopsis aidingensis sp. nov., a halotolerant Actinobacterium, produces new secondary metabolites. Frontiers in Microbiology, 12, 743116.
Nelms, J., Edwards, R. M., Warwick, J., & Fotheringham, I. (1992). Novel mutations in the pheA gene of Escherichia coli K-12 which result in highly feedback inhibition-resistant variants of chorismate mutase/prephenate dehydratase. Applied and Environmental Microbiology, 58, 2592–2598.
Zhou, H., Liao, X., Wang, T., Du, G., & Chen, J. (2010). Enhanced l-phenylalanine biosynthesis by co-expression of pheA(fbr) and aroF(wt). Bioresource Technology, 101, 4151–4156.
Hamano, Y., Nicchu, I., Shimizu, T., Onji, Y., Hiraki, J., & Takagi, H. (2007). epsilon-Poly-l-lysine producer, Streptomyces albulus, has feedback-inhibition resistant aspartokinase. Applied Microbiology and Biotechnology, 76, 873–882.
Isogai, S., Matsushita, T., Imanishi, H., Koonthongkaew, J., Toyokawa, Y., Nishimura, A., Yi, X., Kazlauskas, R., & Takagi, H. (2021). High-level production of lysine in the yeast Saccharomyces cerevisiae by rational design of homocitrate synthase. Applied and Environmental Microbiology, 87, e0060021.
Zhang, J., Xu, M., Ge, X., Zhang, X., Yang, T., Xu, Z., & Rao, Z. (2017). Reengineering of the feedback-inhibition enzyme N-acetyl-l-glutamate kinase to enhance l-arginine production in Corynebacterium crenatum. Journal of Industrial Microbiology and Biotechnology, 44, 271–283.
Wang, T. W., Zhu, H., Ma, X. Y., Zhang, T., Ma, Y. S., & Wei, D. Z. (2006). Mutant library construction in directed molecular evolution: Casting a wider net. Molecular Biotechnology, 34, 55–68.
Reshetnikov, A. S., Khmelenina, V. N., Mustakhimov, I. I., Kalyuzhnaya, M., Lidstrom, M., & Trotsenko, Y. A. (2011). Diversity and phylogeny of the ectoine biosynthesis genes in aerobic, moderately halophilic methylotrophic bacteria. Extremophiles, 15, 653–663.
Harayama, S. (1998). Artificial evolution by DNA shuffling. Trends in Biotechnology, 16, 76–82.
Biot-Pelletier, D., & Martin, V. J. (2014). Evolutionary engineering by genome shuffling. Applied Microbiology and Biotechnology, 98, 3877–3887.
Stephanopoulos, G. (2002). Metabolic engineering by genome shuffling. Nature Biotechnology, 20, 666–668.
Zeng, W., Chen, G., Wu, H., Wang, J., Liu, Y., Guo, Y., & Liang, Z. (2016). Improvement of Bacillus subtilis for poly-gamma-glutamic acid production by genome shuffling. Microbial Biotechnology, 9, 824–833.
Li, S., Chen, X., Dong, C., Zhao, F., Tang, L., & Mao, Z. (2013). Combining genome shuffling and interspecific hybridization among Streptomyces improved epsilon-poly-l-lysine production. Applied Biochemistry and Biotechnology, 169, 338–350.
Li, S., Li, F., Chen, X. S., Wang, L., Xu, J., Tang, L., & Mao, Z. G. (2012). Genome shuffling enhanced epsilon-poly-l-lysine production by improving glucose tolerance of Streptomyces graminearus. Applied Biochemistry and Biotechnology, 166, 414–423.
Cao, X., Hou, L., Lu, M., Wang, C., & Zeng, B. (2010). Genome shuffling of Zygosaccharomyces rouxii to accelerate and enhance the flavour formation of soy sauce. Journal of the Science of Food and Agriculture, 90, 281–285.
Zheng, P., Liu, M., Liu, X. D., Du, Q. Y., Ni, Y., & Sun, Z. H. (2012). Genome shuffling improves thermotolerance and glutamic acid production of Corynebacterium glutamicum. World Journal of Microbiology and Biotechnology, 28, 1035–1043.
Huang, Q. G., Zeng, B. D., Liang, L., Wu, S. G., & Huang, J. Z. (2018). Genome shuffling and high-throughput screening of Brevibacterium flavum MDV1 for enhanced l-valine production. World Journal of Microbiology and Biotechnology, 34, 121.
Wang, L., Chen, X., Wu, G., Zeng, X., Ren, X., Li, S., Tang, L., & Mao, Z. (2016). Genome shuffling and gentamicin-resistance to improve epsilon-poly-l-lysine productivity of Streptomyces albulus W-156. Applied Biochemistry and Biotechnology, 180, 1601–1617.
Webb, B., & Sali, A. (2016). Comparative protein structure modeling using MODELLER. Current Protocols in Protein Science, 86, 2.9.1-2.9.37.
Webb, B., & Sali, A. (2021). Protein structure modeling with MODELLER. Methods in Molecular Biology, 2199, 239–255.
DiMaio, F. (2017). Rosetta structure prediction as a tool for solving difficult molecular replacement problems. Methods in Molecular Biology, 1607, 455–466.
Das, R., & Baker, D. (2008). Macromolecular modeling with Rosetta. Annual Review of Biochemistry, 77, 363–382.
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Zidek, A., Potapenko, A., Bridgland, A., Meyer, C., Kohl, S. A. A., Ballard, A. J., Cowie, A., Romera-Paredes, B., Nikolov, S., Jain, R., Adler, J., … Hassabis, D. (2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596, 583–589.
Cramer, P. (2021). AlphaFold2 and the future of structural biology. Nature Structural and Molecular Biology, 28, 704–705.
Carvalho, H. F., Barbosa, A. J., Roque, A. C., Iranzo, O., & Branco, R. J. (2017). Integration of molecular dynamics based predictions into the optimization of de novo protein designs: Limitations and benefits. Methods in Molecular Biology, 1529, 181–201.
Czech, L., Hoppner, A., Kobus, S., Seubert, A., Riclea, R., Dickschat, J. S., Heider, J., Smits, S. H. J., & Bremer, E. (2019). Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis. Scientific Reports, 9, 364.
Widderich, N., Kobus, S., Hoppner, A., Riclea, R., Seubert, A., Dickschat, J. S., Heider, J., Smits, S. H., & Bremer, E. (2016). Biochemistry and crystal structure of ectoine synthase: A metal-containing member of the cupin superfamily. PLoS ONE, 11, e0151285.
Chen, W., Zhang, S., Jiang, P., Yao, J., He, Y., Chen, L., Gui, X., Dong, Z., & Tang, S. Y. (2015). Design of an ectoine-responsive AraC mutant and its application in metabolic engineering of ectoine biosynthesis. Metabolic Engineering, 30, 149–155.
Salinas, V. H., & Ranganathan, R. (2018). Coevolution-based inference of amino acid interactions underlying protein function. eLife, 7, e34300.
Wu, Z., Liu, H., Xu, L., Chen, H. F., & Feng, Y. (2020). Algorithm-based coevolution network identification reveals key functional residues of the alpha/beta hydrolase subfamilies. The FASEB Journal, 34, 1983–1995.
Bhattacharya, S., Margheritis, E. G., Takahashi, K., Kulesha, A., D’Souza, A., Kim, I., Yoon, J. H., Tame, J. R. H., Volkov, A. N., Makhlynets, O. V., & Korendovych, I. V. (2022). NMR-guided directed evolution. Nature, 610, 389–393.
Mascotti, M. L. (2022). Resurrecting enzymes by ancestral sequence reconstruction. Methods in Molecular Biology, 2397, 111–136.
Cai, W., Pei, J., & Grishin, N. V. (2004). Reconstruction of ancestral protein sequences and its applications. BMC Ecology and Evolution, 4, 33.
Saelens, J. W., Sweeney, M. I., Viswanathan, G., Xet-Mull, A. M., Jurcic Smith, K. L., Sisk, D. M., Hu, D. D., Cronin, R. M., Hughes, E. J., Brewer, W. J., Coers, J., Champion, M. M., Champion, P. A., Lowe, C. B., Smith, C. M., Lee, S., Stout, J. E., & Tobin, D. M. (2022). An ancestral mycobacterial effector promotes dissemination of infection. Cell, 185, 4507-4525.e4518.
Wang, T., Liang, C., Xing, W., Wu, W., Hou, Y., Zhang, L., Xiao, S., Xu, H., An, Y., Zheng, M., Liu, L., & Nie, L. (2020). Transcriptional factor engineering in microbes for industrial biotechnology. Journal of Chemical Technology and Biotechnology, 95, 3071–3078.
Nicoll, C. R., Bailleul, G., Fiorentini, F., Mascotti, M. L., Fraaije, M. W., & Mattevi, A. (2020). Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs. Nature Structural and Molecular Biology, 27, 14–24.
Zhou, H. X., Rivas, G., & Minton, A. P. (2008). Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annual Review of Biophysics, 37, 375–397.
Sanyal, N., Arentson, B. W., Luo, M., Tanner, J. J., & Becker, D. F. (2015). First evidence for substrate channeling between proline catabolic enzymes: A validation of domain fusion analysis for predicting protein–protein interactions. Journal of Biological Chemistry, 290, 2225–2234.
Wang, T., Qin, X., Liang, C., & Yuan, H. (2018). Engineering substrate channeling in biosystems for improved efficiency. Journal of Chemical Technology and Biotechnology, 93, 3364–3373.
Dueber, J. E., Wu, G. C., Malmirchegini, G. R., Moon, T. S., Petzold, C. J., Ullal, A. V., Prather, K. L., & Keasling, J. D. (2009). Synthetic protein scaffolds provide modular control over metabolic flux. Nature Biotechnology, 27, 753–759.
Conrado, R. J., Wu, G. C., Boock, J. T., Xu, H., Chen, S. Y., Lebar, T., Turnsek, J., Tomsic, N., Avbelj, M., Gaber, R., Koprivnjak, T., Mori, J., Glavnik, V., Vovk, I., Bencina, M., Hodnik, V., Anderluh, G., Dueber, J. E., Jerala, R., & DeLisa, M. P. (2012). DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Research, 40, 1879–1889.
Widderich, N., Czech, L., Elling, F. J., Konneke, M., Stoveken, N., Pittelkow, M., Riclea, R., Dickschat, J. S., Heider, J., & Bremer, E. (2016). Strangers in the archaeal world: Osmostress-responsive biosynthesis of ectoine and hydroxyectoine by the marine thaumarchaeon Nitrosopumilus maritimus. Environmental Microbiology, 18, 1227–1248.
Schulz, A., Stoveken, N., Binzen, I. M., Hoffmann, T., Heider, J., & Bremer, E. (2017). Feeding on compatible solutes: A substrate-induced pathway for uptake and catabolism of ectoines and its genetic control by EnuR. Environmental Microbiology, 19, 926–946.
Schulz, A., Hermann, L., Freibert, S. A., Bonig, T., Hoffmann, T., Riclea, R., Dickschat, J. S., Heider, J., & Bremer, E. (2017). Transcriptional regulation of ectoine catabolism in response to multiple metabolic and environmental cues. Environmental Microbiology, 19, 4599–4619.
Sauer, T., & Galinski, E. A. (1998). Bacterial milking: A novel bioprocess for production of compatible solutes. Biotechnology and Bioengineering, 57, 306–313.
Lang, Y. J., Bai, L., Ren, Y. N., Zhang, L. H., & Nagata, S. (2011). Production of ectoine through a combined process that uses both growing and resting cells of Halomonas salina DSM 5928T. Extremophiles, 15, 303–310.
Schubert, T., Maskow, T., Benndorf, D., Harms, H., & Breuer, U. (2007). Continuous synthesis and excretion of the compatible solute ectoine by a transgenic, nonhalophilic bacterium. Applied Environmental Microbiology, 73, 3343–3347.
Ning, Y., Wu, X., Zhang, C., Xu, Q., Chen, N., & Xie, X. (2016). Pathway construction and metabolic engineering for fermentative production of ectoine in Escherichia coli. Metabolic Engineering, 36, 10–18.
Bethlehem, L., & Moritz, K. D. (2020). Boosting Escherichia coli’s heterologous production rate of ectoines by exploiting the non-halophilic gene cluster from Acidiphilium cryptum. Extremophiles, 24, 733–747.
Perez-Garcia, F., Ziert, C., Risse, J. M., & Wendisch, V. F. (2017). Improved fermentative production of the compatible solute ectoine by Corynebacterium glutamicum from glucose and alternative carbon sources. Journal of Biotechnology, 258, 59–68.
Giesselmann, G., Dietrich, D., Jungmann, L., Kohlstedt, M., Jeon, E. J., Yim, S. S., Sommer, F., Zimmer, D., Muhlhaus, T., Schroda, M., Jeong, K. J., Becker, J., & Wittmann, C. (2019). Metabolic engineering of Corynebacterium glutamicum for high-level ectoine production: Design, combinatorial assembly, and implementation of a transcriptionally balanced heterologous ectoine pathway. Biotechnology Journal, 14, e1800417.
Cho, S., Lee, Y. S., Chai, H., Lim, S. E., Na, J. G., & Lee, J. (2022). Enhanced production of ectoine from methane using metabolically engineered Methylomicrobium alcaliphilum 20Z. Biotechnology for Biofuels and Bioproducts, 15, 5.
Cantera, S., Lebrero, R., Rodríguez, S., García-Encina, P. A., & Muñoz, R. (2017). Ectoine bio-milking in methanotrophs: A step further towards methane-based bio-refineries into high added-value products. Chemical Engineering Journal, 328, 44–48.
Cantera, S., Phandanouvong-Lozano, V., Pascual, C., García-Encina, P. A., Lebrero, R., Hay, A., & Muñoz, R. (2020). A systematic comparison of ectoine production from upgraded biogas using Methylomicrobium alcaliphilum and a mixed haloalkaliphilic consortium. Waste Management, 102, 773–781.
Zhang, S., Fang, Y., Zhu, L., Li, H., Wang, Z., Li, Y., & Wang, X. (2021). Metabolic engineering of Escherichia coli for efficient ectoine production. Systems Microbiology and Biomanufacturing, 1, 444–458.
Fatollahi, P., Ghasemi, M., Yazdian, F., & Sadeghi, A. (2021). Ectoine production in bioreactor by Halomonas elongata DSM2581: Using MWCNT and Fe-nanoparticle. Biotechnology Progress, 37, e3073.
Chen, W.-C., Yuan, F.-W., Wang, L.-F., Chien, C.-C., & Wei, Y.-H. (2020). Ectoine production with indigenous Marinococcus sp. MAR2 isolated from the marine environment. Preparative Biochemistry and Biotechnology, 50, 74–81.
Czech, L., Stoveken, N., & Bremer, E. (2016). EctD-mediated biotransformation of the chemical chaperone ectoine into hydroxyectoine and its mechanosensitive channel-independent excretion. Microbial Cell Factories, 15, 126.
Acknowledgements
The publication of this paper was financially supported by a Fund from Anhui Polytechnic University (2022YQQ063). We apologize for authors whose publications relevant to this paper's topic were not cited for various reasons.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest in this publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, W., Liu, K., Kong, F. et al. Multiple Functions of Compatible Solute Ectoine and Strategies for Constructing Overproducers for Biobased Production. Mol Biotechnol (2023). https://doi.org/10.1007/s12033-023-00827-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-023-00827-7