Abstract
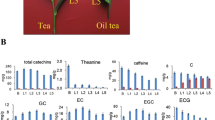
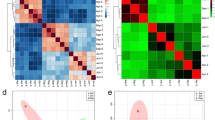
This study aimed to explore the molecular mechanisms underlying the differential quality of tea made from leaves at different development stages. Fresh Camellia sinensis (L.) O. Kuntze “Sichuan Colonial” leaves of various development stages, from buds to old leaves, were subjected to transcriptome sequencing and metabolome analysis, and the DESeq package was used for differential expression analysis, followed by functional enrichment analyses and protein interaction analysis. Target metabolome analysis indicated that the contents of most compounds, including theobromine and epicatechin gallate, were lowest in old leaves, and transcriptome analysis revealed that DEGs were significantly involved in extracellular regions and phenylpropanoid biosynthesis, photosynthesis-related pathways, and the oleuropein steroid biosynthesis pathway. Protein–protein interaction analysis identified LOC114256852 as a hub gene. Caffeine, theobromine, l-theanine, and catechins were the main metabolites of the tea leaves, and the contents of all four main metabolites were the lowest in old leaves. Phenylpropanoid biosynthesis, photosynthesis, and brassinosteroid biosynthesis may be important targets for breeding efforts to improve tea quality.






Similar content being viewed by others
References
Zhou, C., Mei, X., Rothenberg, D. O. N., Yang, Z., Zhang, W., Wan, S., Yang, H., & Zhang, L. (2020). Metabolome and transcriptome analysis reveals putative genes involved in Anthocyanin accumulation and coloration in white and pink tea (Camellia sinensis) flower. Molecules, 25, 190.
Zhao, L., Chen, C., Wang, Y., Shen, J., & Ding, Z. (2019). Conserved microRNA act boldly during sprout development and quality formation in Pingyang Tezaocha (Camellia sinensis). Frontiers in genetics, 10, 237.
Liu, S., An, Y., Li, F., Li, S., Liu, L., Zhou, Q., Zhao, S., & Wei, C. (2018). Genome-wide identification of simple sequence repeats and development of polymorphic SSR markers for genetic studies in tea plant (Camellia sinensis). Molecular Breeding, 38, 1–13.
Rothenberg, D. O. N., Zhou, C., & Zhang, L. (2018). A review on the weight-loss effects of oxidized tea polyphenols. Molecules, 23, 1176.
Rothenberg, D. O. N., & Zhang, L. (2019). Mechanisms underlying the anti-depressive effects of regular tea consumption. Nutrients, 11, 1361.
Fang, W.-P., Meinhardt, L. W., Tan, H.-W., Zhou, L., Mischke, S., & Zhang, D. (2014). Varietal identification of tea (Camellia sinensis) using nanofluidic array of single nucleotide polymorphism (SNP) markers. Horticulture research, 1, 1–8.
Guo, F., Guo, Y., Wang, P., Wang, Y., & Ni, D. (2017). Transcriptional profiling of catechins biosynthesis genes during tea plant leaf development. Planta, 246, 1139–1152.
Ashihara, H., Deng, W. W., Mullen, W., & Crozier, A. (2010). Distribution and biosynthesis of flavan-3-ols in Camellia sinensis seedlings and expression of genes encoding biosynthetic enzymes. Phytochemistry, 71, 559–566.
Eungwanichayapant, P., & Popluechai, S. (2009). Accumulation of catechins in tea in relation to accumulation of mRNA from genes involved in catechin biosynthesis. Plant Physiology and Biochemistry, 47, 94–97.
Wu, H., Chen, D., Li, J., Yu, B., Qiao, X., Huang, H., & He, Y. (2013). De novo characterization of leaf transcriptome using 454 sequencing and development of EST-SSR markers in tea (Camellia sinensisCamellia sinensis). Plant Molecular Biology Reporter, 2013(31), 524–538.
Wu, Z.-J., Li, X.-H., Liu, Z.-W., Xu, Z.-S., & Zhuang, J. (2014). De novo assembly and transcriptome characterization: Novel insights into catechins biosynthesis in Camellia sinensis. BMC Plant Biology, 14, 1–16.
Li, F.-D., Tong, W., Xia, E.-H., & Wei, C.-L. (2019). Optimized sequencing depth and de novo assembler for deeply reconstructing the transcriptome of the tea plant, an economically important plant species. BMC Bioinformatics, 20, 1–11.
Wen, G. (2017). A simple process of RNA-sequence analyses by Hisat2, Htseq and DESeq2. In Proceedings of the 2017 international conference on biomedical engineering and bioinformatics.
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., Salzberg, S. L., Wold, B. J., & Pachter, L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology, 28, 511–515.
Anders, S., & Huber, W. (2012). Differential expression of RNA-Seq data at the gene level–the DESeq package. European Molecular Biology Laboratory (EMBL).
Kolde, R., & Kolde, M. R. (2015). Package ‘pheatmap.’ R package (vol. 1, p. 790).
Alexa, A., Rahnenfuhrer, J., Alexa, M. A. and Suggests, A. (2016). Package ‘topGO’.
Jin, J., Tian, F., Yang, D.-C., Meng, Y.-Q., Kong, L., Luo, J., & Gao, G. (2016). PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research, 45(D1), D1040–D1045.
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., Simonovic, M., Roth, A., Santos, A., & Tsafou, K. P. (2015). STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Research, 43(D1), D447–D452.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., & Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13, 2498–2504.
Ninomiya, M., Unten, L., & Kim, M. (1997). Chemical and physicochemical properties of green tea polyphenols. In T. Yamamoto, L. R. Juneja, D. C. Chu, & M. Kim (Eds.), Chemistry and applications of green tea (pp. 23–35). CRC Press.
Jin, J., Zhou, C., Ma, C., Yao, M., Ma, J., & Chen, L. (2014). Identification on purine alkaloids of representative tea germplasms in China. Journal of Plant Genetic Resources 2014(15), 285.
Xia, E.-H., Zhang, H.-B., Sheng, J., Li, K., Zhang, Q.-J., Kim, C., Zhang, Y., Liu, Y., Zhu, T., & Li, W. (2017). The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Molecular Plant, 10, 866–877.
Feng, L., Gao, M.-J., Hou, R.-Y., Hu, X.-Y., Zhang, L., Wan, X.-C., & Wei, S. (2014). Determination of quality constituents in the young leaves of albino tea cultivars. Food Chemistry, 155, 98–104.
Yang, C., Hu, Z., Lu, M., Li, P., Tan, J., Chen, M., Lv, H., Zhu, Y., Zhang, Y., & Guo, L. (2018). Application of metabolomics profiling in the analysis of metabolites and taste quality in different subtypes of white tea. Food Research International, 106, 909–919.
Du, G.-J., Zhang, Z., Wen, X.-D., Yu, C., Calway, T., Yuan, C.-S., & Wang, C.-Z. (2012). Epigallocatechin gallate (EGCG) is the most effective cancer chemopreventive polyphenol in green tea. Nutrients, 4, 1679–1691.
Tounekti, T., Joubert, E., Hernández, I., & Munné-Bosch, S. (2013). Improving the polyphenol content of tea. Critical Reviews in Plant Sciences, 32, 192–215.
Lin, Y.-S., Tsai, Y.-J., Tsay, J.-S., & Lin, J.-K. (2003). Factors affecting the levels of tea polyphenols and caffeine in tea leaves. Journal of Agricultural and Food Chemistry, 51, 1864–1873.
Park, J.-S., Kim, J.-B., Hahn, B.-S., Kim, K.-H., Ha, S.-H., Kim, J.-B., & Kim, Y.-H. (2004). EST analysis of genes involved in secondary metabolism in Camellia sinensis (tea), using suppression subtractive hybridization. Plant Science, 166, 953–961.
George, V. C., Dellaire, G., & Rupasinghe, H. V. (2017). Plant flavonoids in cancer chemoprevention: Role in genome stability. The Journal of Nutritional Biochemistry, 45, 1–14.
Wang, T.-Y., Li, Q., & Bi, K.-S. (2018). Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian Journal of Pharmaceutical Sciences, 13, 12–23.
Li, X., Zhang, L., Ahammed, G. J., Li, Z.-X., Wei, J.-P., Shen, C., Yan, P., Zhang, L.-P., & Han, W.-Y. (2017). Nitric oxide mediates brassinosteroid-induced flavonoid biosynthesis in Camellia sinensis L. Journal of Plant Physiology, 214, 145–151.
Li, X., Zhang, L.-P., Zhang, L., Yan, P., Ahammed, G. J., & Han, W.-Y. (2019). Methyl salicylate enhances flavonoid biosynthesis in tea leaves by stimulating the phenylpropanoid pathway. Molecules, 24, 362.
Li, X., Zhang, L., Ahammed, G. J., Li, Z.-X., Wei, J.-P., Shen, C., Yan, P., Zhang, L.-P., & Han, W.-Y. (2017). Stimulation in primary and secondary metabolism by elevated carbon dioxide alters green tea quality in Camellia sinensis L. Scientific Reports, 7, 1–12.
Li, Z.-X., Yang, W.-J., Ahammed, G. J., Shen, C., Yan, P., Li, X., & Han, W.-Y. (2016). Developmental changes in carbon and nitrogen metabolism affect tea quality in different leaf position. Plant Physiology and Biochemistry, 106, 327–335.
Zhou, H., Xu, M., Pan, H., & Yu, X. (2015). Leaf-age effects on temperature responses of photosynthesis and respiration of an alpine oak, Quercus aquifolioides, in southwestern China. Tree Physiology, 35, 1236–1248.
Planas-Riverola, A., Gupta, A., Betegón-Putze, I., Bosch, N., Ibañes, M., & Caño-Delgado, A. I. (2019). Brassinosteroid signaling in plant development and adaptation to stress. Development, 146(5), dev151894.
Jiang, Y.-P., Cheng, F., Zhou, Y.-H., Xia, X.-J., Mao, W.-H., Shi, K., Chen, Z.-X., & Yu, J.-Q. (2012). Hydrogen peroxide functions as a secondary messenger for brassinosteroids-induced CO2 assimilation and carbohydrate metabolism in Cucumis sativus. Journal of Zhejiang University Science B, 13, 811–823.
Li, X., Ahammed, G. J., Li, Z.-X., Zhang, L., Wei, J.-P., Shen, C., Yan, P., Zhang, L.-P., & Han, W.-Y. (2016). Brassinosteroids improve quality of summer tea (Camellia sinensis L.) by balancing biosynthesis of polyphenols and amino acids. Frontiers in Plant Science, 7, 1304.
Funding
This study was supported by Key R&D Program of Science and Technology in Tibet Autonomous Region, China (Grant No. XZ202001ZY0035N), Tibet Key Research and Development Program (Grant No. XZ202001ZY004).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Zh., Zhang, Gq., Zhang, Zw. et al. Target Metabolome and Transcriptome Analysis Reveal Molecular Mechanism Associated with Changes of Tea Quality at Different Development Stages. Mol Biotechnol 65, 52–60 (2023). https://doi.org/10.1007/s12033-022-00525-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00525-w