Abstract
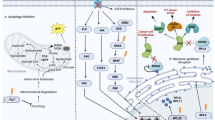
Oncogenic metabolic reprogramming impacts the abundance of key metabolites that regulate signaling and epigenetics. Metabolic vulnerability in the cancer cell is evident from the Warburg effect. The research on metabolism in the progression and survival of breast cancer (BC) is under focus. Oncogenic signal activation and loss of tumor suppressor are important regulators of tumor cell metabolism. Several intrinsic and extrinsic factors contribute to metabolic reprogramming. The molecular mechanisms underpinning metabolic reprogramming in BC are extensive and only partially defined. Various signaling pathways involved in the metabolism play a significant role in the modulation of BC. Notably, PI3K/AKT/mTOR pathway, lactate-ERK/STAT3 signaling, loss of the tumor suppressor Ras, Myc, oxidative stress, activation of the cellular hypoxic response and acidosis contribute to different metabolic reprogramming phenotypes linked to enhanced glycolysis. The alterations in mitochondrial genes have also been elaborated upon along with their functional implications. The outcome of these active research areas might contribute to the development of novel therapeutic interventions and the remodeling of known drugs.





Similar content being viewed by others
References
Momenimovahed Z, Salehiniya H. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer: Targets and Therapy. 2019;11:151.
Sung, H., J. Ferlay, R.L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, et al., Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians, 2021. 71(3): p. 209–249.
Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin. 2011;61:409–18.
Sung H, Ferlay J, Siegel RL. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Łukasiewicz S, Czeczelewski M, Forma A. Breast cancer-epidemiology, risk factors, classification, prognostic markers, and current treatment strategies-an updated review. Cancers. 2021;13(17):4287.
Vazquez A, Kamphorst JJ, Markert EK, Schug ZT, Tardito S, Gottlieb E. Cancer metabolism at a glance. J Cell Sci. 2016;129(18):3367–73.
Landor SK-J, Mutvei AP, Mamaeva V, Jin S, Busk M, Borra R, et al. Hypo-and hyperactivated Notch signaling induce a glycolytic switch through distinct mechanisms. Proc Natl Acad Sci. 2011;108(46):18814–9.
Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–69.
Anderson G. Type I diabetes pathoetiology and pathophysiology: roles of the gut microbiome, pancreatic cellular interactions, and the ‘bystander’ activation of memory CD8+ T cells. Int J Mol Sci. 2023;24(4):3300.
Formentini L, Martínez-Reyes I, Cuezva JM. The mitochondrial bioenergetic capacity of carcinomas. IUBMB Life. 2010. https://doi.org/10.1002/iub.352.
Yadav UP, Singh T, Kumar P, Sharma P, Kaur H, Sharma S, et al. Metabolic adaptations in cancer stem cells. Front Oncol. 2020;10:1010.
Zheng J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation. Oncol Lett. 2012;4(6):1151–7.
Végran F, Boidot R, Michiels C, Sonveaux P, Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-κB/IL-8 pathway that drives tumor angiogenesis. Can Res. 2011;71(7):2550–60.
Rothberg, J.M., K.M. Bailey, J.W. Wojtkowiak, Y. Ben-Nun, M. Bogyo, E. Weber, et al., Acid-mediated tumor proteolysis: contribution of cysteine cathepsins. Neoplasia, 2013. 15(10): p. 1125-IN9.
Hu Y, Lu W, Chen G, Wang P, Chen Z, Zhou Y, et al. K-rasG12V transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res. 2012;22(2):399–412.
Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–37.
Schiliro C, Firestein BL. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. 2021;10(5):1056.
Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17(11):1498–503.
King, A., M. Selak, and, and E. Gottlieb, Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene, 2006. 25(34): p. 4675–4682.
Albadari, N., S. Deng, and W. Li, The transcriptional factors HIF-1 and HIF-2 and their novel inhibitors in cancer therapy. 2019. 14(7): p. 667–682.
Laurenti G, Tennant DA. Isocitrate dehydrogenase (IDH), succinate dehydrogenase (SDH), fumarate hydratase (FH): three players for one phenotype in cancer? Biochem Soc Trans. 2016;44(4):1111–6.
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30.
Tarrado-Castellarnau M, de Atauri P, Cascante M. Oncogenic regulation of tumor metabolic reprogramming. Oncotarget. 2016;7(38):62726–53.
Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer Nature Reviews Disease Primers. 2019;5(1):66.
Dewan K, Mandal A. Surrogate molecular classification of breast carcinoma: A classification in need or a dilemma indeed. Oncology Journal of India. 2020;4(3):79–86.
Wang Z, Jiang Q, Dong C. Metabolic reprogramming in triple-negative breast cancer. Cancer Biol Med. 2020;17(1):44–59.
Holloway, R.W. and P.A. Marignani, Targeting mTOR and Glycolysis in HER2-Positive Breast Cancer. Cancers (Basel), 2021. 13(12), 2922.
Cappelletti V, Iorio E, Miodini P, Silvestri M, Dugo M, Daidone MG. Metabolic footprints and molecular subtypes in breast cancer. Dis Markers. 2017;2017:7687851.
Shin E, Koo JS. Glucose metabolism and glucose transporters in breast cancer. Front Cell Dev Biol. 2021;9: 728759.
Long J-P, Li X-N, Zhang F. Targeting metabolism in breast cancer: How far we can go? World journal of clinical oncology. 2016;7(1):122.
Choi J, Jung W-H, Koo JS. Metabolism-related proteins are differentially expressed according to the molecular subtype of invasive breast cancer defined by surrogate immunohistochemistry. Pathobiology. 2013;80(1):41–52.
Zhang, Y., Q. Li, Z. Huang, and B. Li, Targeting glucose metabolism enzymes in cancer treatment: current and emerging strategies. Cancers. 2022. 14(19), 4568.
Ko Y-H, Lin Z, Flomenberg N, Pestell RG, Howell A, Sotgia F, et al. Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells: implications for preventing chemotherapy resistance. Cancer Biol Ther. 2011;12(12):1085–97.
Ko B-H, Paik J-Y, Jung K-H, Lee K-H. 17β-Estradiol augments 18F-FDG uptake and glycolysis of T47D breast cancer cells via membrane-initiated rapid PI3K–Akt activation. J Nucl Med. 2010;51(11):1740–7.
Fhu CW, Ali A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules. 2020;25(17):3935.
Iqbal MA, Bamezai RN. Resveratrol inhibits cancer cell metabolism by down regulating pyruvate kinase M2 via inhibition of mammalian target of rapamycin. PLoS ONE. 2012;7(5): e36764.
Luo S, Jiang Y, Anfu Z, Zhao Y, Wu X, Li M, et al. Targeting hypoxia-inducible factors for breast cancer therapy: A narrative review. Front Pharmacol. 2022;13:1064661.
Miricescu D, Totan A, Stanescu-Spinu I-I, Badoiu SC, Stefani C, Greabu M. PI3K/AKT/mTOR signaling pathway in breast cancer: From molecular landscape to clinical aspects. Int J Mol Sci. 2020;22(1):173.
Kulkoyluoglu-Cotul E, Arca A, Madak-Erdogan Z. Crosstalk between estrogen signaling and breast cancer metabolism. Trends Endocrinol Metab. 2019;30(1):25–38.
Lim, W., B. Mayer, and T. Pawson, Cell signaling. 2014: Taylor & Francis.
Bhaskar PT, Nogueira V, Patra KC, Jeon S-M, Park Y, Robey RB, et al. mTORC1 hyperactivity inhibits serum deprivation-induced apoptosis via increased hexokinase II and GLUT1 expression, sustained Mcl-1 expression, and glycogen synthase kinase 3β inhibition. Mol Cell Biol. 2009;29(18):5136–47.
Pande M, Bondy ML, Do K-A, Sahin AA, Ying J, Mills GB, et al. Association between germline single nucleotide polymorphisms in the PI3K-AKT-mTOR pathway, obesity, and breast cancer disease-free survival. Breast Cancer Res Treat. 2014;147(2):381–7.
Miller TW, Rexer BN, Garrett JT, Arteaga CL. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011;13(6):1–12.
Sobral-Leite M, Salomon I, Opdam M, Kruger DT, Beelen KJ, van der Noort V, et al. Cancer-immune interactions in ER-positive breast cancers: PI3K pathway alterations and tumor-infiltrating lymphocytes. Breast Cancer Res. 2019;21(1):1–12.
Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, et al. Regulation of hypoxia-inducible factor 1α expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22(20):7004–14.
Roberts D, Miyamoto S. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ. 2015;22(2):248–57.
Brown RS, Wahl RL. Overexpression of glut-1 glucose transporter in human breast cancer an immunohistochemical study. Cancer. 1993;72(10):2979–85.
Gaude E, Frezza C. Defects in mitochondrial metabolism and cancer. Cancer Metabolism. 2014;2(1):1–9.
Liu W-S, Chan S-H, Chang H-T, Li G-C, Tu Y-T, Tseng H-H, et al. Isocitrate dehydrogenase 1–snail axis dysfunction significantly correlates with breast cancer prognosis and regulates cell invasion ability. Breast Cancer Res. 2018;20(1):1–17.
Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–62.
Yang L, Hou Y, Yuan J, Tang S, Zhang H, Zhu Q, et al. Twist promotes reprogramming of glucose metabolism in breast cancer cells through PI3K/AKT and p53 signaling pathways. Oncotarget. 2015;6(28):25755.
Kotowski K, Rosik J, Machaj F, Supplitt S, Wiczew D, Jabłońska K, et al. Role of PFKFB3 and PFKFB4 in cancer: genetic basis, impact on disease development/progression, and potential as therapeutic targets. Cancers. 2021;13(4):909.
Liu Y, Wang R, Zhang L, Li J, Lou K, Shi B. The lipid metabolism gene FTO influences breast cancer cell energy metabolism via the PI3K/AKT signaling pathway. Oncol Lett. 2017;13(6):4685–90.
Tseng C-W, Kuo W-H, Chan S-H, Chan H-L, Chang K-J, Wang L-H. Transketolase regulates the metabolic switch to control breast cancer cell metastasis via the α-ketoglutarate signaling pathway. Can Res. 2018;78(11):2799–812.
Miao P, Sheng S, Sun X, Liu J, Huang G. Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life. 2013;65(11):904–10.
He W, Miao FJ-P, Lin DC-H, Schwandner RT, Wang Z, Gao J, et al. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature. 2004;429(6988):188–93.
Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci. 2011;108(49):19611–6.
Mu X, Shi W, Xu Y, Xu C, Zhao T, Geng B, et al. Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle. 2018;17(4):428–38.
Zaugg, M., P.-H. Lou, E. Lucchinetti, M. Gandhi, and A.S. Clanachan, Postconditioning with Intralipid emulsion protects against reperfusion injury in post-infarct remodeled rat hearts by activation of ROS-Akt/Erk signaling. Translational Research, 2017. 186: p. 36–51. e2.
Tkach, M., C. Rosemblit, M.A. Rivas, C.J. Proietti, M.C. Díaz Flaqué, M.F. Mercogliano, et al., p42/p44 MAPK-mediated Stat3Ser727 phosphorylation is required for progestin-induced full activation of Stat3 and breast cancer growth. Endocr Relat Cancer, 2013. 20(2): p. 197–212.
Bromberg J. Stat proteins and oncogenesis. J Clin Investig. 2002;109(9):1139–42.
Epling-Burnette P, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Investig. 2001;107(3):351–62.
Li R, Hebert JD, Lee TA, Xing H, Boussommier-Calleja A, Hynes RO, et al. Macrophage-secreted TNFα and TGFβ1 influence migration speed and persistence of cancer cells in 3D tissue culture via independent pathways. Can Res. 2017;77(2):279–90.
Yao A, Xiang Y, Si YR, Fan LJ, Li JP, Li H, et al. PKM2 promotes glucose metabolism through a let-7a-5p/Stat3/hnRNP-A1 regulatory feedback loop in breast cancer cells. J Cell Biochem. 2019;120(4):6542–54.
David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463(7279):364–8.
Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149(6):1269–83.
Wang L, Zhang S, Wang X. The metabolic mechanisms of breast cancer metastasis. Front Oncol. 2021;10: 602416.
Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci. 2011;108(39):16259–64.
Hsu C-C, Tseng L-M, Lee H-C. Role of mitochondrial dysfunction in cancer progression. Exp Biol Med. 2016;241(12):1281–95.
Karlsson, R., E. Pedersen, Z. Wang, and C. Brakebusch, Rho GTPase function in tumorigenesis. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 2009. 1796(2): p. 91–98.
Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4(1):1–15.
Turgeon M-O, Perry NJ, Poulogiannis G. DNA damage, repair, and cancer metabolism. Front Oncol. 2018;8:15.
Maréchal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5(9): a012716.
Aird KM, Worth AJ, Snyder NW, Lee JV, Sivanand S, Liu Q, et al. ATM couples replication stress and metabolic reprogramming during cellular senescence. Cell Rep. 2015;11(6):893–901.
Turgeon M-O, Perry NJS, Poulogiannis G. DNA damage, repair, and cancer metabolism. Front Oncol. 2018. https://doi.org/10.3389/fonc.2018.00015.
Biswas DK, Iglehart JD. Linkage between EGFR family receptors and nuclear factor kappaB (NF-κB) signaling in breast cancer. J Cell Physiol. 2006;209(3):645–52.
Shi J, Wei PK. Interleukin-8: A potent promoter of angiogenesis in gastric cancer. Oncol Lett. 2016;11(2):1043–50.
Heidemann J, Ogawa H, Dwinell MB, Rafiee P, Maaser C, Gockel HR, et al. Angiogenic effects of interleukin 8 (CXCL8) in human intestinal microvascular endothelial cells are mediated by CXCR2*. J Biol Chem. 2003;278(10):8508–15.
Rizzo, M., L. Varnier, G. Pezzicoli, M. Pirovano, L. Cosmai, and C. Porta, IL-8 and its role as a potential biomarker of resistance to anti-angiogenic agents and immune checkpoint inhibitors in metastatic renal cell carcinoma. Frontiers in Oncology, 2022: p. 12, 4411.
Sudarshan S, Sourbier C, Kong H-S, Block K, Romero VV, Yang Y, et al. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and HIF-1α stabilization by glucose-dependent generation of reactive oxygen species. Mol Cell Biol. 2009;29(15):4080.
Shanmugasundaram K, Nayak B, Shim E-H, Livi CB, Block K, Sudarshan S. The oncometabolite fumarate promotes pseudohypoxia through noncanonical activation of NF-κB signaling. J Biol Chem. 2014;289(35):24691–9.
Korherr C, Gille H, Schäfer R, Koenig-Hoffmann K, Dixelius J, Egland KA, et al. Identification of proangiogenic genes and pathways by high-throughput functional genomics: TBK1 and the IRF3 pathway. Proc Natl Acad Sci. 2006;103(11):4240–5.
Kamdje AHN, Etet PFS, Vecchio L, Muller JM, Krampera M, Lukong KE. Signaling pathways in breast cancer: therapeutic targeting of the microenvironment. Cell Signal. 2014;26(12):2843–56.
Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12(8):445–64.
Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294(5545):1337–40.
Funasaka T, Hogan V, Raz A. Phosphoglucose isomerase/autocrine motility factor mediates epithelial and mesenchymal phenotype conversions in breast cancer. Can Res. 2009;69(13):5349–56.
Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83.
Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci. 2004;101(10):3329–35.
Andrade-Vieira R, Xu Z, Colp P, Marignani PA. Loss of LKB1 expression reduces the latency of ErbB2-mediated mammary gland tumorigenesis, promoting changes in metabolic pathways. PLoS ONE. 2013;8(2): e56567.
Zhang Z-G, Zhang H-S, Sun H-L, Liu H-Y, Liu M-Y, Zhou Z. KDM5B promotes breast cancer cell proliferation and migration via AMPK-mediated lipid metabolism reprogramming. Exp Cell Res. 2019;379(2):182–90.
Boland M, Chourasia A, Macleod K. Mitochondrial Dysfunction in Cancer. Front Oncol. 2013. https://doi.org/10.3389/fonc.2013.00292.
Lamb R, Ablett MP, Spence K, Landberg G, Sims AH, Clarke RB. Wnt pathway activity in breast cancer sub-types and stem-like cells. PLoS ONE. 2013;8(7): e67811.
Jeng K-S, Sheen I-S, Jeng W-J, Yu M-C, Hsiau H-I, Chang F-Y. High expression of Sonic Hedgehog signaling pathway genes indicates a risk of recurrence of breast carcinoma. Onco Targets Ther. 2014;7:79.
Hu J, Li T, Du S, Chen Y, Wang S, Xiong F, et al. The MAPK signaling pathway mediates the GPR91-dependent release of VEGF from RGC-5 cells. Int J Mol Med. 2015;36(1):130–8.
Huang Q, Cao H, Zhan L, Sun X, Wang G, Li J, et al. Mitochondrial fission forms a positive feedback loop with cytosolic calcium signaling pathway to promote autophagy in hepatocellular carcinoma cells. Cancer Lett. 2017;403:108–18.
Xie Q, Wu Q, Horbinski CM, Flavahan WA, Yang K, Zhou W, et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nat Neurosci. 2015;18(4):501–10.
Ma Y, Wang L, Jia R. The role of mitochondrial dynamics in human cancers. Am J Cancer Res. 2020;10(5):1278.
Bushel PR, Ward J, Burkholder A, Li J, Anchang B. Mitochondrial-nuclear epistasis underlying phenotypic variation in breast cancer pathology. Sci Rep. 2022;12(1):1393.
Kim TW, Kim B, Kim JH, Kang S, Park S-B, Jeong G, et al. Nuclear-encoded mitochondrial MTO1 and MRPL41 are regulated in an opposite epigenetic mode based on estrogen receptor status in breast cancer. BMC Cancer. 2013;13(1):502.
Chang S, Singh L, Thaker K, Abedi S, Singh MK, Patel TH, et al. Altered retrograde signaling patterns in breast cancer cells cybrids with H and J mitochondrial DNA haplogroups. Int J Mol Sci. 2022;23(12):6687.
El-Sahli S, Wang L. Cancer stem cell-associated pathways in the metabolic reprogramming of breast cancer. Int J Mol Sci. 2020;21(23):9125.
Oku Y, Nishiya N, Shito T, Yamamoto R, Yamamoto Y, Oyama C, et al. Small molecules inhibiting the nuclear localization of YAP/TAZ for chemotherapeutics and chemosensitizers against breast cancers. FEBS Open Bio. 2015;5:542–9.
Ma J, Fan Z, Tang Q, Xia H, Zhang T, Bi F. Aspirin attenuates YAP and β-catenin expression by promoting β-TrCP to overcome docetaxel and vinorelbine resistance in triple-negative breast cancer. Cell Death Dis. 2020;11(7):530.
Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16(4):357–66.
Ishikawa T, Hosaka YZ, Beckwitt C, Wells A, Oltvai ZN, Warita K. Concomitant attenuation of HMG-CoA reductase expression potentiates the cancer cell growth-inhibitory effect of statins and expands their efficacy in tumor cells with epithelial characteristics. Oncotarget. 2018;9(50):29304.
Wang P, Gong Y, Guo T, Li M, Fang L, Yin S, et al. Activation of Aurora A kinase increases YAP stability via blockage of autophagy. Cell Death Dis. 2019;10(6):432.
Turkson J, Zhang S, Mora LB, Burns A, Sebti S, Jove R. A novel platinum compound inhibits constitutive Stat3 signaling and induces cell cycle arrest and apoptosis of malignant cells. J Biol Chem. 2005;280(38):32979–88.
Kim JW, Gautam J, Kim JE, Kim J, Kang KW. Inhibition of tumor growth and angiogenesis of tamoxifen-resistant breast cancer cells by ruxolitinib, a selective JAK2 inhibitor. Oncol Lett. 2019;17(4):3981–9.
Chen K-F, Tai W-T, Hsu C-Y, Huang J-W, Liu C-Y, Chen P-J, et al. Blockade of STAT3 activation by sorafenib derivatives through enhancing SHP-1 phosphatase activity. Eur J Med Chem. 2012;55:220–7.
Srirangam A, Milani M, Mitra R, Guo Z, Rodriguez M, Kathuria H, et al. The human immunodeficiency virus protease inhibitor ritonavir inhibits lung cancer cells, in part, by inhibition of survivin. J Thorac Oncol. 2011;6(4):661–70.
Park S, Chang C-Y, Safi R, Liu X, Baldi R, Jasper JS, et al. ERRα-regulated lactate metabolism contributes to resistance to targeted therapies in breast cancer. Cell Rep. 2016;15(2):323–35.
Wu T, Harder BG, Wong PK, Lang JE, Zhang DD. Oxidative stress, mammospheres and Nrf2–new implication for breast cancer therapy? Mol Carcinog. 2015;54(11):1494–502.
Park S, Safi R, Liu X, Baldi R, Liu W, Liu J, et al. Inhibition of ERRα prevents mitochondrial pyruvate uptake exposing NADPH-generating pathways as targetable vulnerabilities in breast cancer. Cell Rep. 2019;27(12):3587.
Feldinger K, Kong A. Profile of neratinib and its potential in the treatment of breast cancer. Breast Cancer: Targets and Therapy. 2015;7:147.
Barry JB, Giguere V. Epidermal growth factor–induced signaling in breast cancer cells results in selective target gene activation by orphan nuclear receptor estrogen-related receptor α. Can Res. 2005;65(14):6120–9.
Madan B, Ke Z, Harmston N, Ho SY, Frois A, Alam J, et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. 2016;35(17):2197–207.
Furuya K, Sasaki A, Tsunoda Y, Tsuji M, Udaka Y, Oyamada H, et al. Eribulin upregulates miR-195 expression and downregulates Wnt3a expression in non-basal-like type of triple-negative breast cancer cell MDA-MB-231. Hum Cell. 2016;29:76–82.
Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci. 2013;110(50):20224–9.
Tam BY, Chiu K, Chung H, Bossard C, Nguyen JD, Creger E, et al. The CLK inhibitor SM08502 induces anti-tumor activity and reduces Wnt pathway gene expression in gastrointestinal cancer models. Cancer Lett. 2020;473:186–97.
Katoh M. Antibody-drug conjugate targeting protein tyrosine kinase 7, a receptor tyrosine kinase-like molecule involved in WNT and vascular endothelial growth factor signaling: effects on cancer stem cells, tumor microenvironment and whole-body homeostasis. Annals Transl Med. 2017;5(23):462.
Fischer MM, Cancilla B, Yeung VP, Cattaruzza F, Chartier C, Murriel CL, et al. WNT antagonists exhibit unique combinatorial antitumor activity with taxanes by potentiating mitotic cell death. Sci Adv. 2017;3(6): e1700090.
Grzybowska-Szatkowska L, Ślaska B. Mitochondrial NADH dehydrogenase polymorphisms are associated with breast cancer in Poland. J Appl Genet. 2014;55(2):173–81.
Czarnecka AM, Klemba A, Krawczyk T, Zdrozny M, Arnold RS, Bartnik E, et al. Mitochondrial NADH-dehydrogenase polymorphisms as sporadic breast cancer risk factor. Oncol Rep. 2010;23(2):531–5.
Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Can Res. 2005;65(17):8028–33.
Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai R. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007;249(2):249–55.
Czarnecka AM, Krawczyk T, Plak K, Klemba A, Zdrozny M, Arnold RS, et al. Mitochondrial genotype and breast cancer predisposition. Oncol Rep. 2010;24(6):1521–34.
Tan D-J, Bai R-K, Wong L-JC. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Can Res. 2002;62(4):972–6.
Gallardo ME, Moreno-Loshuertos R, López C, Casqueiro M, Silva J, Bonilla F, et al. m 6267G> A: a recurrent mutation in the human mitochondrial DNA that reduces cytochrome c oxidase activity and is associated with tumors. Hum Mutat. 2006;27(6):575–82.
Girolimetti G, Marchio L, De Leo A, Mangiarelli M, Amato LB, Zanotti S, et al. Mitochondrial DNA analysis efficiently contributes to the identification of metastatic contralateral breast cancers. J Cancer Res Clin Oncol. 2021;147(2):507–16.
Grzybowska-Szatkowska L, Ślaska B, Rzymowska J, Brzozowska A, Floriańczyk B. Novel mitochondrial mutations in the ATP6 and ATP8 genes in patients with breast cancer. Mol Med Rep. 2014;10(4):1772–8.
Kalia M. Personalized oncology: Recent advances and future challenges. Metabolism. 2013;62:S11–4.
Jayasekera LP, Ranasinghe R, Senathilake KS, Kotelawala JT, de Silva K, Abeygunasekara PH, et al. Mitochondrial genome in sporadic breast cancer: A case control study and a proteomic analysis in a Sinhalese cohort from Sri Lanka. PLoS ONE. 2023;18(2): e0281620.
Jackson SE, Chester JD. Personalised cancer medicine. Int J Cancer. 2015;137(2):262–6.
Gambardella V, Tarazona N, Cejalvo JM, Lombardi P, Huerta M, Roselló S, et al. Personalized medicine: recent progress in cancer therapy. Cancers. 2020;12(4):1009.
Manzari MT, Shamay Y, Kiguchi H, Rosen N, Scaltriti M, Heller DA. Targeted drug delivery strategies for precision medicines. Nat Rev Mater. 2021;6(4):351–70.
Acknowledgements
We acknowledge the Central University of Punjab for providing infrastructure and facilities, University Grant Commission, New Delhi, for Junior Research Fellowship (JRF) to Ruthuparna Malayil for PhD, Council of Scientific and Industrial Research (CSIR), New Delhi, for Senior Research Fellowship (SRF) to Yogita Chhichholiya and Tashvinder Singh, and Science and Engineering Research Board (SERB) to Harsh Vikram Singh for Project Assistantship. DST-FIST grant (SR/FST/LS-I/2017/49) to the Department of Human Genetics and Molecular Medicine, Central University of Punjab, is acknowledged with thanks.
Funding
Financial support to Rhuthuparna Malayil (Award No- KL1216200313) from University Grant Commission, India, Yogita Chhichholiya (Award No- 09/1051(0038)/2019-EMR-1) and Tashvinder Singh (Award No- 09/1051(0042)/2019-EMR-1) from Council of Scientific and Industrial Research (CSIR) India, and Harsh Vikram Singh (SRG/2021/001806) for Project Assistantship from Science and Engineering Research Board (SERB) is highly acknowledged.
Author information
Authors and Affiliations
Contributions
Anjana Munshi and Sandeep Singh conceptualized the idea of the review. Rhuthuparna malayil, Yogita Chhichholiya, Kanika Vasudeva, Harsh Vikram Singh, and Tashvinder Singh curated the data and prepared the draft. Anjana Munshi and Sandeep Singh critically revised and edited the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Malayil, R., Chhichholiya, Y., Vasudeva, K. et al. Oncogenic metabolic reprogramming in breast cancer: focus on signaling pathways and mitochondrial genes. Med Oncol 40, 174 (2023). https://doi.org/10.1007/s12032-023-02037-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-023-02037-2