Abstract
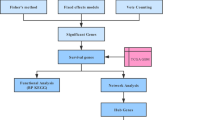
Glioblastoma (GBM) is the most aggressive intracranial tumors. Despite the comprehensive treatments, the median survival of GBM patients is still dismal. Consequently, it is critical to explore potential biomarkers and underlying molecular mechanisms of GBM. We integrated two datasets (GSE19728 and GSE 4290) to identify differentially expressed genes (DEGs) of GBM. Eighty-two GBM samples and 31 brain normal samples were screened by using GEO2R and Draw Venn Diagram. We carried out Database for Annotation, Visualization and Integrated Discovery (DAVID) to analyze gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) to analyze the DEGs. To further screen hub genes, protein–protein interaction (PPI) of these DEGs was visualized by Cytoscape with Search Tool for the Retrieval of Interacting Genes (STRING). GEPIA and UALCAN website were utilized to identify the hub genes expression and survival data. In total, 568 common DEGs were determined, including 141 upregulated genes and 427 downregulated genes. Thirty-five hub genes were identified in the highest module consisting of 35 nodes and 535 edges, which are mainly associated with cell cycle, p53 signaling pathway. According to the further analysis results of hub genes, we found that the PDZ-binding kinase (PBK) gene had a high expression and significantly worse survival in GBM contrasted to brain normal samples (P < 0.05). PBK could be a potential prognostic factor and therapeutic target for GBM treatment. In the future, the potential biomarkers for significant prognostic information can be preliminarily assessed using this method, although further experimentations are needed to verify the results.






Similar content being viewed by others
References
Abe Y, Matsumoto S, Kito K, Ueda N (2000) Cloning and expression of a novel MAPKK-like protein kinase, lymphokine-activated killer T-cell-originated protein kinase, specifically expressed in the testis and activated lymphoid cells. J Biol Chem 275(28):21525–21531
Bo L, Wei B, Li C, Wang Z, Gao Z, Miao Z (2017) Identification of potential key genes associated with glioblastoma based on the gene expression profile. Oncol Lett 14:2045–2052
Brown-Clay JD, Shenoy DN, Timofeeva O, Kallakury BV, Nandi AK, Banerjee PP (2015) PBK/TOPK enhances aggressive phenotype in prostate cancer via beta-catenin-TCF/LEF-mediated matrix metalloproteinases production and invasion. Oncotarget 6(17):15594–15609
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19(8):649–658
Chang CF, Chen SL, Sung WW, Hsieh MJ, Hsu HT, Chen LH, Chen MK, Ko JL, Chen CJ, Chou MC (2016) PBK/TOPK expression predicts prognosis in oral cancer. Int J Mol Sci 17:7
Cheng W, Ren X, Zhang C, Cai J, Liu Y, Han S, Wu A (2016) Bioinformatic profiling identifies an immune-related risk signature for glioblastoma. Neurology 86(24):2226–2234
Davis S, Meltzer PS (2007) GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23(14):1846–1847
Dou X, Wei J, Sun A, Shao G, Childress C, Yang W, Lin Q (2015) PBK/TOPK mediates geranylgeranylation signaling for breast cancer cell proliferation. Cancer Cell Int 15:27
Ferrer VP, Moura Neto V, Mentlein R (2018) Glioma infiltration and extracellular matrix: key players and modulators. Glia 66(8):1542–1565
Furth N, Aylon Y, Oren M (2018) p53 shades of Hippo. Cell Death Differ 25(1):81–92
Gaudet S, Branton D, Lue RA (2000) Characterization of PDZ-binding kinase, a mitotic kinase. Proc Natl Acad Sci U S A 97(10):5167–5172
Gene Ontology C (2006) The Gene Ontology (GO) project in 2006. Nucleic Acids Res 34(Database issue):D322–D326
Hsu FH, Serpedin E, Hsiao TH, Bishop AJ, Dougherty ER, Chen Y (2012) Reducing confounding and suppression effects in TCGA data: an integrated analysis of chemotherapy response in ovarian cancer. BMC Genomics 13(Suppl 6):S13
Hu F, Gartenhaus RB, Eichberg D, Liu Z, Fang HB, Rapoport AP (2010) PBK/TOPK interacts with the DBD domain of tumor suppressor p53 and modulates expression of transcriptional targets including p21. Oncogene 29(40):5464–5474
Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57
Joel M, Mughal AA, Grieg Z, Murrell W, Palmero S, Mikkelsen B, Fjerdingstad HB, Sandberg CJ, Behnan J, Glover JC, Langmoen IA, Stangeland B (2015) Targeting PBK/TOPK decreases growth and survival of glioma initiating cells in vitro and attenuates tumor growth in vivo. Mol Cancer 14:121
Kawakami A, Fisher DE (2016) Bioinformatic analysis of gene expression for melanoma treatment. J Invest Dermatol 136(12):2342–2344
Kwon CH, Park HJ, Choi YR, Kim A, Kim HW, Choi JH, Hwang CS, Lee SJ, Choi CI, Jeon TY, Kim DH, Kim GH, Park do Y (2016) PSMB8 and PBK as potential gastric cancer subtype-specific biomarkers associated with prognosis. Oncotarget 7(16):21454–21468
Le Large TY, Mato Prado M, Krell J, Bijlsma MF, Meijer LL, Kazemier G, Frampton AE, Giovannetti E (2016) Bioinformatic analysis reveals pancreatic cancer molecular subtypes specific to the tumor and the microenvironment. Expert Rev Mol Diagn 16(7):733–736
Lei B1, Liu S, Qi W, Zhao Y, Li Y, Lin N, Xu X, Zhi C, Mei J, Yan Z, Wan L, Shen H (2013) PBK/TOPK expression in non-small-cell lung cancer: its correlation and prognostic significance with Ki67 and p53 expression. Histopathology 63(5):696–703
Li VD, Li KH, Li JT (2019) TP53 mutations as potential prognostic markers for specific cancers: analysis of data from the Cancer Genome Atlas and the International Agency for Research on Cancer TP53 Database. J Cancer Res Clin Oncol 145(3):625–636
Liu Y, Liu H, Cao H, Song B, Zhang W, Zhang W (2015) PBK/TOPK mediates promyelocyte proliferation via Nrf2-regulated cell cycle progression and apoptosis. Oncol Rep 34(6):3288–3296
Liu Y, Li F, Yang YT, Xu XD, Chen JS, Chen TL, Chen HJ, Zhu YB, Lin JY, Li Y, Xie XM, Sun XL, Ke YQ (2019) IGFBP2 promotes vasculogenic mimicry formation via regulating CD144 and MMP2 expression in glioma. Oncogene 38(11):1815–1831
Ohashi T, Komatsu S, Ichikawa D, Miyamae M, Okajima W, Imamura T, Kiuchi J, Nishibeppu K, Kosuga T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E (2016) Overexpression of PBK/TOPK contributes to tumor development and poor outcome of esophageal squamous cell carcinoma. Anticancer Res 36(12):6457–6466
Park JH, Lin ML, Nishidate T, Nakamura Y, Katagiri T (2006) PDZ-binding kinase/T-LAK cell-originated protein kinase, a putative cancer/testis antigen with an oncogenic activity in breast cancer. Cancer Res 66(18):9186–9195
Qian X, Tan H, Zhang J, Liu K, Yang T, Wang M, Debinskie W, Zhao W, Chan MD, Zhou X (2016) Identification of biomarkers for pseudo and true progression of GBM based on radiogenomics study. Oncotarget 7(34):55377–55394
Roth P, Silginer M, Goodman SL, Hasenbach K, Thies S, Maurer G, Schraml P, Tabatabai G, Moch H, Tritschler I, Weller M (2013) Integrin control of the transforming growth factor-beta pathway in glioblastoma. Brain 136(Pt 2):564–576
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Song BN, Kim SK, Chu IS (2017) Bioinformatic identification of prognostic signature defined by copy number alteration and expression of CCNE1 in non-muscle invasive bladder cancer. Exp Mol Med 49(1):e282
Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO, European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups, National Cancer Institute of Canada Clinical Trials Group (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10(5):459–466
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45(D1):D362–D368
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45(W1):W98–W102
Tanimura A, Yamazaki M, Hashimotodani Y, Uchigashima M, Kawata S, Abe M, Kita Y, Hashimoto K, Shimizu T, Watanabe M, Sakimura K, Kano M (2010) The endocannabinoid 2-arachidonoylglycerol produced by diacylglycerol lipase alpha mediates retrograde suppression of synaptic transmission. Neuron 65(3):320–327
Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38(Web Server issue:W214–W220
Xing Z, Chu C, Chen L, Kong X (2016) The use of Gene Ontology terms and KEGG pathways for analysis and prediction of oncogenes. Biochim Biophys Acta 1860(11 Pt B):2725–2734
Xu M, Xu S (2019) PBK/TOPK overexpression and survival in solid tumors: a PRISMA-compliant meta-analysis. Medicine (Baltimore) 98(10):e14766
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dong, C., Fan, W. & Fang, S. PBK as a Potential Biomarker Associated with Prognosis of Glioblastoma. J Mol Neurosci 70, 56–64 (2020). https://doi.org/10.1007/s12031-019-01400-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-019-01400-1