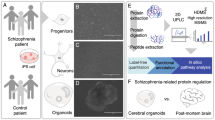
Abstract
Human induced pluripotent stem cells (hiPSCs) have been employed very successfully to identify molecular and cellular features of psychiatric disorders that would be impossible to discover in traditional postmortem studies. Despite the wealth of new available information though, there is still a critical need to establish quantifiable and accessible molecular markers that can be used to reveal the biological causality of the disease. In this paper, we introduce a new quantitative framework based on supervised learning to investigate structural alterations in the neuronal cytoskeleton of hiPSCs of schizophrenia (SCZ) patients. We show that, by using Support Vector Machines or selected Artificial Neural Networks trained on image-based features associated with somas of hiPSCs derived neurons, we can predict very reliably SCZ and healthy control cells. In addition, our method reveals that \(\beta\)III tubulin and FGF12, two critical components of the cytoskeleton, are differentially regulated in SCZ and healthy control cells, upon perturbation by GSK3 inhibition.



Similar content being viewed by others
References
Anguera, M. S., Sadreyev, R., Zhang, Z., Szanto, A., Payer, B., Sheridan, S. D., Kwok, S., Haggarty, S. J., Sur, M., Alvarez, J., et al. (2012). Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell, 11(1), 75–90.
Balan, S., Toyoshima, M., & Yoshikawa, T. (2019). Contribution of induced pluripotent stem cell technologies to the understanding of cellular phenotypes in schizophrenia. Neurobiology of disease, 131, 104162.
Bauer, D. E., Haroutunian, V., McCullumsmith, R. E., & Meador-Woodruff, J. H. (2009). Expression of four housekeeping proteins in elderly patients with schizophrenia. Journal of Neural Transmission, 116(4), 487–491.
Benítez-King, G., Valdés-Tovar, M., Trueta, C., Galvan-Arrieta, T., Argueta, J., Alarcon, S., et al. (2016). The microtubular cytoskeleton of olfactory neurons derived from patients with schizophrenia or with bipolar disorder: Implications for biomarker characterization, neuronal physiology and pharmacological screening. Molecular and Cellular Neuroscience, 73, 84–95.
Bentea, E., Depasquale, E. A. K., O’Donovan, S. M., Sullivan, C. R., Simmons, M., Meador-Woodruff, J. H., Zhou, Y., Xu, C., Bai, B., Peng, J., et al. (2019). Kinase network dysregulation in a human induced pluripotent stem cell model of disc1 schizophrenia. Molecular omics, 15(3), 173–188.
Brennand, K., Savas, J. N., Kim, Y., Tran, N., Simone, A., Hashimoto-Torii, K., Beaumont, K. G., Kim, H. J., Topol, A., Ladran, I., et al. Phenotypic differences in hipsc npcs derived from patients with schizophrenia. Molecular Psychiatry, 20(3), 361–368.
Brennand, K. J., Simone, A., Jou, J., Gelboin-Burkhart, C., Tran, N., Sangar, S., Li, Y., Mu, Y., Chen, G., Yu, D., et al. (2011). Modelling schizophrenia using human induced pluripotent stem cells. Nature, 473(7346), 221–225.
Chadha, R., Alganem, K., Mccullumsmith, R. E., & Meador-Woodruff, J. H. (2021). Mtor kinase activity disrupts a phosphorylation signaling network in schizophrenia brain. Molecular Psychiatry, pp. 1–12.
Pyradiomics Community. (2016). pyradiomics.readthedocs.io/en/latest/index.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., et al. (2020) An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929.
Habela, C. W., Song, H., & Ming, G-L. (2016). Modeling synaptogenesis in schizophrenia and autism using human ipsc derived neurons. Molecular and Cellular Neuroscience, 73, 52–62.
Haralick, R. M., Shanmugam, K., & Dinstein, I. (1973). Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics, SMC–3.
Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2012). Imagenet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems, 25, 1097–1105.
LeCun, Y., Bottou, L., Bengio, Y., & Haffner, P. (1998). Gradient-based learning applied to document recognition. Proceedings of the IEEE, 86(11), 2278–2324.
Li, J. J., & Tong, X. (2020). Statistical hypothesis testing versus machine learning binary classification: Distinctions and guidelines. Patterns, 1(7), 100115.
Lin, W., Hasenstab, K., Cunha, G. M., & Schwartzman, A. (2020). Comparison of handcrafted features and convolutional neural networks for liver mr image adequacy assessment. Scientific Reports, 10(1), 1–11.
Marchisella, F., Coffey, E. T., & Hollos, P. (2016). Microtubule and microtubule associated protein anomalies in psychiatric disease. Cytoskeleton, 73(10), 596–611.
McGuire, J. L., Depasquale, E. A., Funk, A. J., O’Donnovan, S. M., Hasselfeld, K., Marwaha, S., Hammond, J. H., Hartounian, V., Meador-Woodruff, J. H., Meller, J., et al. (2017). Abnormalities of signal transduction networks in chronic schizophrenia. NPJ schizophrenia, 3(1), 1–10.
Moehle, M. S., Luduena, R. F., Haroutunian, V., Meador-Woodruff, J. H., & McCullumsmith, R. E. (2012). Regional differences in expression of \(\beta\)-tubulin isoforms in schizophrenia. Schizophrenia Research, 135(1-3), 181–186.
Neyshabur, B., Li, Z., Bhojanapalli, S., LeCun, Y., & Srebro, N. (2018). The role of over-parametrization in generalization of neural networks. In International Conference on Learning Representations.
O’Shea, K., & Nash, R. (2015). An introduction to convolutional neural networks. arXiv preprint arXiv:1511.08458.
Park, I.-H., Arora, N., Huo, H., Maherali, N., Ahfeldt, T., Shimamura, A., et al. (2008). Disease-specific induced pluripotent stem cells. Cell, 134(5), 877–886.
Rodrigues-Amorim, D., Rivera-Baltanás, T., Vallejo-Curto, M. d. C., Rodriguez-Jamardo, C., de Las Heras, E., Barreiro-Villar, C., Blanco-Formoso, M., Fernández-Palleiro, P., Álvarez-Ariza, M., López, M., et al. (2020) Plasma \(\beta\)-iii tubulin, neurofilament light chain and glial fibrillary acidic protein are associated with neurodegeneration and progression in schizophrenia. Scientific Reports, 10(1), 1–10.
Safari, K., Prasad, S., & Labate, D. (2020). A multiscale deep learning approach for high-resolution hyperspectral image classification. IEEE Geoscience and Remote Sensing Letters.
Smith, L. N., & Topin, N. (2019). Super-convergence: very fast training of neural networks using large learning rates. In Tien Pham (Ed.), Artificial Intelligence and Machine Learning for Multi-Domain Operations Applications, vol. 11006 (pp. 369–386). SPIE: International Society for Optics and Photonics.
Stertz, L., Di Re, J., Pei, G., Fries, G. R., Mendez, E., Li, S., Smith-Callahan, L., Raventos, H., Tipo, J., Cherukuru, R., et al. (2021). Convergent genomic and pharmacological evidence of pi3k/gsk3 signaling alterations in neurons from schizophrenia patients. Neuropsychopharmacology, 46(3), 673–682.
Sun, C., & Wee, W. G. (1983). Neighboring gray level dependence matrix for texture classification. Computer Vision, Graphics, and Image Processing, 23(3), 341–352.
Thibault, G., Angulo, J., & Meyer, F. (2014). Advanced statistical matrices for texture characterization: application to cell classification. IEEE Transactions of Biomedical Imaging, 61(3), 630–7.
Tiihonen, J., Taipale, H., Mehtälä, J., Vattulainen, P., Correll, C. U., & Tanskanen, A. (2019). Association of antipsychotic polypharmacy vs monotherapy with psychiatric rehospitalization among adults with schizophrenia. JAMA Psychiatry, 76(5), 499–507.
Van Griethuysen, J. J. M., Fedorov, A., Parmar, C., Hosny, A., Aucoin, N., Narayan, V., Beets-Tan, R. G. H., Fillion-Robin, J. -C., Pieper, S., & Aerts, H. J. W. L. (2017). Computational radiomics system to decode the radiographic phenotype. Cancer research, 77(21), e104–e107.
Vapnik, V. N. (1999). An overview of statistical learning theory. IEEE Transactions on Neural Networks, 10(5), 988–999.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., Kaiser, L., & Polosukhin, I. (2017). Attention is all you need. In Advances in neural information processing systems, pages 5998–6008.
Wen, Z., Nguyen, H. N., Guo, Z., Lalli, M. A., Wang, X., Su, Y., Kim, N. -S., Yoon, K. -J., Shin, J., Zhang, C., et al. (2014). Synaptic dysregulation in a human ips cell model of mental disorders. Nature, 515(7527), 414–418.
Wightman, R. (2019). Pytorch image models. https://github.com/rwightman/pytorch-image-models.
Yu, D. X., Di Giorgio, F. P., Yao, J., Marchetto, M. C., Brennand, K., Wright, R., Mei, A., Mchenry, L., Lisuk, D., Grasmick, J. M., et al. (2014). Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports, 2(3), 295–310.
Zwanenburg, A., Vallières, M., Abdalah, M. A., Aerts, H. J. W. L., Andrearczyk, V., Apte, A., Ashrafinia, S., Bakas, S., Beukinga, R. J., Boellaard, R., et al. (2020). The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology, 295(2), 328–338.
Acknowledgements
NF and DL acknowledge financial support by NSF-DMS 1720487 and 1720452; they also acknowledge the Hewlett Packard Enterprise Data Science Institute at the University of Houston for providing computational resources. FL was supported by 1R01MH124351 and JDR was supported by a National Institutes of Health training grant (T32ES007254).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fularczyk, N., Di Re, J., Stertz, L. et al. A Learning Based Framework for Disease Prediction from Images of Human-Derived Pluripotent Stem Cells of Schizophrenia Patients. Neuroinform 20, 513–523 (2022). https://doi.org/10.1007/s12021-022-09561-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-022-09561-y