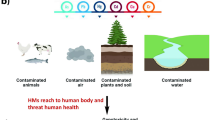
Abstract
Cadmium (Cd) is one of the most well-known toxic metals capable of entering the human body via the food chain, leading to serious health problems. Human gut microbes play a pivotal role in controlling Cd bioavailability and toxicity within the human gastrointestinal tract, primarily due to their capacity for Cd adsorption and metabolism. In this work, a Cd-resistant bacterial strain, Enterococcus faecalis strain ATCC19433 was isolated from human gut microbiota. Cd binding assays and comprehensive characterization analyses were performed, revealing the ability of strain ATCC19433 to remove Cd from the solution. Cd adsorption primarily occurred on the bacterial cell walls, which was ascribed to the exciting of functional groups on the bacterial surfaces, containing alkyl, amide II, and phosphate groups; meanwhile, Cd could enter cells, probably through transport channels or via diffusion. These results indicated that Cd removal by the strain was predominantly dependent on biosorption and bioaccumulation. Whole-genome sequencing analyses further suggested the probable mechanisms of biosorption and bioaccumulation, including Cd transport by transporter proteins, active efflux of Cd by cadmium efflux pumps, and mitigating oxidative stress-induced cell damage by DNA repair proteases. This study evaluated the Cd removal capability and mechanism of Enterococcus faecalis strain ATCC19433 while annotating the genetic functions related to Cd removal, which may facilitate the development of potential human gut strains for the removal of Cd.








Similar content being viewed by others
Data Availability
All the data obtained in this study are presented in this paper, and the specific sequencing results and element determination data can be obtained from the author (yuzheng330@126.com).
References
Turner A (2019) Cadmium pigments in consumer products and their health risks. Sci Total Environ 657:1409–1418
Hu L, Zhong H, He Z (2021) Toxicity evaluation of cadmium-containing quantum dots: a review of optimizing physicochemical properties to diminish toxicity. Colloids Surf B Biointerfaces 200:111609
Huttunen-Saarivirta E, Korpiniemi H, Kuokkala VT, Paajanen H (2013) Corrosion of cadmium plating by runway de-icing chemicals: study of surface phenomena and comparison of corrosion tests. Surf Coat Technol 232:101–115
Zhuang Z, Nino-Savala AG, Mi Z-d, Wan Y-n, Su D-c, Li H-f, Fangmeier A (2021) Cadmium accumulation in wheat and maize grains from China: interaction of soil properties, novel enrichment models and soil thresholds. Environ Pollut 275:116623
Duan Z, Zheng Y, Luo Y, Wu Y, Wen J, Wu J (2021) Evaluation of cadmium transfer from soil to the human body through maize consumption in a cadmium anomaly area of southwestern China. Environ Toxicol Chem 40:2923–2934
Xu M, Yang L, Chen Y, Jing H, Wu P, Yang W (2022) Selection of rice and maize varieties with low cadmium accumulation and derivation of soil environmental thresholds in karst. Ecotoxicol Environ Saf 247:114244
Rinaldi M, Micali A, Marini H, Adamo EB, Puzzolo D, Pisani A, Trichilo V, Altavilla D, Squadrito F, Minutoli L (2017) Cadmium, organ toxicity and therapeutic approaches: a review on brain, kidney and testis damage. Curr Med Chem 24:3879–3893
Andjelkovic M, Djordjevic AB, Antonijevic E, Antonijevic B, Stanic M, Kotur-Stevuljevic J, Spasojevic-Kalimanovska V, Jovanovic M, Boricic N, Wallace D, Bulat Z (2019) Toxic effect of acute cadmium and lead exposure in rat blood, liver, and kidney. Int J Environ Res Public Health 16(2):274
Thursby E, Juge N (2017) Introduction to the human gut microbiota. Biochem J 474:1823–1836
Yin N, Zhao Y, Wang P, Du H, Yang M, Han Z, Chen X, Sun G, Cui Y (2021) Effect of gut microbiota on in vitro bioaccessibility of heavy metals and human health risk assessment from ingestion of contaminated soils*. Environ Pollut 279:116943
Wang L, Yin X, Gao S, Jiang T, Ma C (2020) In vitro oral bioaccessibility investigation and human health risk assessment of heavy metals in wheat grains grown near the mines in North China. Chemosphere 252:126522
Duan H, Yu L, Tian F, Zhai Q, Fan L, Chen W (2020) Gut microbiota: a target for heavy metal toxicity and a probiotic protective strategy. Sci Total Environ 742:140429
Arun KB, Madhavan A, Sindhu R, Emmanual S, Binod P, Pugazhendhi A, Sirohi R, Reshmy R, Awasthi MK, Gnansounou E, Pandey A (2021) Probiotics and gut microbiome-prospects and challenges in remediating heavy metal toxicity. J Hazard Mater 420:126676
Zhai Q, Guo Y, Tang X, Tian F, Zhao J, Zhang H, Chen W (2019) Removal of cadmium from rice by Lactobacillus plantarum fermentation. Food Control 96:357–364
Teem H, Seppo S, Jussi M, Raija T, Kalle L (2008) Reversible surface binding of cadmium and lead by lactic acid and bifidobacteria. Int J Food Microbiol 125:170–175
Motta-Romero HA, Perez-Donado CE, Auchtung JM, Rose DJ (2023) Toxicity of cadmium on dynamic human gut microbiome cultures and the protective effect of cadmium-tolerant bacteria autochthonous to the gut. Chemosphere 338:139581
Wang X, Li D, Gao P, Gu W, He X, Yang W, Tang W (2020) Analysis of biosorption and biotransformation mechanism of Pseudomonas chengduensis strain MBR under Cd(II) stress from genomic perspective. Ecotoxicol Environ Saf 198:110655
Panthee S, Paudel A, Hamamoto H, Ogasawara AA, Iwasa T, Blom J, Sekimizu K (2021) Complete genome sequence and comparative genomic analysis of Enterococcus faecalis EF-2001, a probiotic bacterium. Genomics 113:1534–1542
Nueno-Palop C, Narbad A (2011) Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int J Food Microbiol 145:390–394
Chen Y-r, Fang S-t, Liu H-y, Zheng H-m, He Y, Chen Z-w, Chen M-x, Zhang G-x, Zhou H-w (2018) Degradation of trimethylamine in vitro and in vivo by Enterococcus faecalis isolated from healthy human gut. Int Biodeterior Biodegradation 135:24–32
Buttimer C, Sutton T, Colom J, Murray E, Bettio PH, Smith L, Bolocan AS, Shkoporov A, Oka A, Liu B, Herzog JW, Sartor RB, Draper LA, Ross RP, Hill C (2022) Impact of a phage cocktail targeting Escherichia coli and Enterococcus faecalis as members of a gut bacterial consortium in vitro and in vivo. Front. microbiol 13:936083
Halttunen T, Collado MC, El-Nezami H, Meriluoto J, Salminen S (2008) Combining strains of lactic acid bacteria may reduce their toxin and heavy metal removal efficiency from aqueous solution. Lett Appl Microbiol 46:160–165
Ren Y-y, Zhu Z-y, Dong F-y, Song Q-y (2018) Screening, characteristics and mechanism of Cd-tolerance Cunninghamella bertholletiae. J Clean Prod 191:480–489
Zhao H, Zhou F, Qi Y, Dziugan P, Bai F, Walczak P, Zhang B (2013) Screening of Lactobacillus strains for their ability to bind Benzo(a)pyrene and the mechanism of the process. Food Chem Toxicol 59:67–71
Li W, Chen Y, Wang T (2021) Cadmium biosorption by lactic acid bacteria Weissella viridescens ZY-6. Food Control 123:107747
Shen L, Chen R, Wang J, Fan L, Cui L, Zhang Y, Cheng J, Wu X, Li J, Zeng W (2021) Biosorption behavior and mechanism of cadmium from aqueous solutions by Synechocystis sp. PCC6803. RSC Advances 11:18637–18650
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
Ofomaja AE, Ho Y-S (2007) Effect of pH on cadmium biosorption by coconut copra meal. J Hazard Mater 139:356–362
Hlihor RM, Diaconu M, Leon F, Curteanu S, Tavares T, Gavrilescu M (2015) Experimental analysis and mathematical prediction of Cd(II) removal by biosorption using support vector machines and genetic algorithms. New Biotechnology 32:358–368
Mishra B, Boyanov M, Bunker BA, Kelly SD, Kemner KM, Fein JB (2010) High- and low-affinity binding sites for Cd on the bacterial cell walls of Bacillus subtilis and Shewanella oneidensis. Geochim Cosmochim Acta 74:4219–4233
Yin R, Zhai Q, Yu L, Xiao Y, Wang G, Yu R, Tian F, Chen W (2016) The binding characters study of lead removal by Lactobacillus plantarum CCFM8661. Eur Food Res Technol 242:1621–1629
McEldowney (2000) The impact of surface attachment on cadmium accumulation by Pseudomonas fluorescens H2. FEMS Microbiol Ecol 33:121–128
Richter J, Ploderer M, Mongelard G, Gutierrez L, Hauser M-T (2017) Role of CrRLK1L cell wall sensors HERCULES1 and 2, THESEUS1, and FERONIA in growth adaptation triggered by heavy metals and trace elements. Front Plant Sci 8:01554
Chapot-Chartier M-P, Kulakauskas S (2014) Cell wall structure and function in lactic acid bacteria. Microb Cell Fact 13(Suppl 1):S9
Yang Y, Pei J (2020) Isolation and characterization of an Enterococcus strain from Chinese sauerkraut with potential for lead removal. Eur Food Res Technol 246:2055–2064
Massadeh AM, Al-Momani FA, Haddad HI (2005) Removal of lead and cadmium by halophilic bacteria isolated from the Dead Sea shore, Jordan. Biol Trace Elem Res 108:259–269
Huang H, Jia Q, Jing W, Dahms H-U, Wang L (2020) Screening strains for microbial biosorption technology of cadmium. Chemosphere 251:126428
Xia L, Xu X, Zhu W, Huang Q, Chen W (2015) A comparative study on the biosorption of Cd2+ onto Paecilomyces lilacinus XLA and Mucoromycote sp XLC. Int J Mol Sci 16:15670–15687
Chakravarty R, Banerjee PC (2012) Mechanism of cadmium binding on the cell wall of an acidophilic bacterium. Bioresour Technol 108:176–183
Sheng Y, Wang Y, Yang X, Zhang B, He X, Xu W, Huang K (2016) Cadmium tolerant characteristic of a newly isolated Lactococcus lactis subsp lactis. Environ Toxicol Pharmacol 48:183–190
Li P-S, Tao H-C (2015) Cell surface engineering of microorganisms towards adsorption of heavy metals. Crit Rev Microbiol 41:140–149
Gola D, Dey P, Bhattacharya A, Mishra A, Malik A, Namburath M, Ahammad SZ (2016) Multiple heavy metal removal using an entomopathogenic fungi Beauveria bassiana. Bioresour Technol 218:388–396
Pereira M, Bartolome MC, Sanchez-Fortun S (2013) Bioadsorption and bioaccumulation of chromium trivalent in Cr(III)-tolerant microalgae: a mechanisms for chromium resistance. Chemosphere 93:1057–1063
Zhai Q, Tian F, Wang G, Zhao J, Liu X, Cross K, Zhang H, Narbad A, Chen W (2016) The cadmium binding characteristics of a lactic acid bacterium in aqueous solutions and its application for removal of cadmium from fruit and vegetable juices. RSC Adv 6:5990–5998
Limcharoensuk T, Sooksawat N, Sumarnrote A, Awutpet T, Kruatrachue M, Pokethitiyook P, Auesukaree C (2015) Bioaccumulation and biosorption of Cd2+ and Zn2+ by bacteria isolated from a zinc mine in Thailand. Ecotoxicol Environ Saf 122:322–330
Zhang F-D, Xu C-H, Li M-Y, Huang A-M, Sun S-Q (2014) Rapid identification of Pterocarpus santalinus and Dalbergia louvelii by FTIR and 2D correlation IR spectroscopy. J Mol Struct 1069:89–95
Li J, Tang C, Zhang M, Fan C, Guo D, An Q, Wang G, Xu H, Li Y, Zhang W, Chen X, Zhao R (2021) Exploring the Cr(VI) removal mechanism of Sporosarcina saromensis M52 from a genomic perspective. Ecotoxicol Environ Saf 225:112767
Wei X, Fang L, Cai P, Huang Q, Chen H, Liang W, Rong X (2011) Influence of extracellular polymeric substances (EPS) on Cd adsorption by bacteria. Environ Pollut 159:1369–1374
Chen J, Wang L, Li W, Zheng X, Li X (2022) Genomic insights into cadmium resistance of a newly isolated, plasmid-free Cellulomonas sp. strain Y8. Front Microbiol 12:784575
Dyla M, Kjaergaard M, Poulsen H, Nissen P (2020) Structure and mechanism of P-type ATPase ion pumps. Annu Rev Biochem 89:583–603
Smith KF, Bibb LA, Schmitt MP, Oram DM (2009) Regulation and activity of a zinc uptake regulator, Zur, in Corynebacterium diphtheriae. J Bacteriol 191:1595–1603
Chien C-C, Huang C-H, Lin Y-W (2013) Characterization of a heavy metal translocating P-type ATPase gene from an environmental heavy metal resistance Enterobacter sp isolate. Appl Biochem Biotechnol 169:1837–1846
Gupta D, Satpati S, Dixit A, Ranjan R (2019) Fabrication of biobeads expressing heavy metal-binding protein for removal of heavy metal from wastewater. Appl Microbiol Biotechnol 103:5411–5420
Shah S, Damare S (2019) Proteomic response of marine-derived Staphylococcus cohnii #NIOSBK35 to varying Cr(vi) concentrations. Metallomics 11:1465–1471
Ogura K, Kaji D, Sasaki M, Otsuka Y, Takemoto N, Miyoshi-Akiyama T, Kikuchi K (2022) Predominance of ST8 and CC1/spa-t1784 methicillin-resistant Staphylococcus aureus isolates in Japan and their genomic characteristics. J Glob Antimicrob Resist 28:195–202
Mori S, Nomura K, Fujikawa K, Osawa T, Shionyu M, Yoda T, Shirai T, Tsuda S, Yoshizawa-Kumagaye K, Masuda S, Nishio H, Yoshiya T, Suzuki S, Muramoto M, Nishiyama K-i, Shimamoto K (2022) Intermolecular interactions between a membrane protein and a glycolipid essential for membrane protein integration. ACS Chem Biol 17:609–618
Mandal SK, Nayak SG, Kanaujia SP (2021) Identification and characterization of metal uptake ABC transporters in Mycobacterium tuberculosis unveil their ligand specificity. Int J Biol Macromol 185:324–337
Neville SL, Sjoehamn J, Watts JA, MacDermott-Opeskin H, Fairweather SJ, Ganio K, Hulyer AC, McGrath AP, Hayes AJ, Malcolm TR, Davies MR, Nomura N, Iwata S, O'Mara ML, Maher MJ, McDevitt CA (2021) The structural basis of bacterial manganese import. Sci Adv 7:32
Cellier MF, Bergevin I, Boyer E, Richer E (2001) Polyphyletic origins of bacterial Nramp transporters. Trends in Genetics17:365–370
Xia X, Wu S, Zhou Z, Wang G (2021) Microbial Cd(II) and Cr(VI) resistance mechanisms and application in bioremediation. J Hazard Mater 401:123685
Roosa S, Wattiez R, Prygiel E, Lesven L, Billon G, Gillan DC (2014) Bacterial metal resistance genes and metal bioavailability in contaminated sediments. Environ Pollut 189:143–151
Liu J, Qu W, Kadiiska MB (2009) Role of oxidative stress in cadmium toxicity and carcinogenesis. Toxicol Appl Pharmacol 238:209–214
Mendoza-Cozatl D, Loza-Tavera H, Hernandez-Navarro A, Moreno-Sanchez R (2005) Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants. FEMS Microbiol Rev 29:653–671
Lazarova N, Krumova E, Stefanova T, Georgieva N, Angelova M (2014) The oxidative stress response of the filamentous yeast Trichosporon cutaneum R57 to copper, cadmium and chromium exposure. Biotechnol Biotechnol Equip 28:855–862
Joe M-H, Jung S-W, Im S-H, Lim S-Y, Song H-P, Kwon O, Kim D-H (2011) Genome-wide response of deinococcus radiodurans on cadmium toxicity. J Microbiol Biotechn 21:438–447
Alav I, Sutton JM, Rahman KM (2018) Role of bacterial efflux pumps in biofilm formation. J Antimicrob Chemother 73:2003–2020
Zhu T-T, Tian L-J, Yu S-S, Yu H-Q (2021) Roles of cation efflux pump in biomineralization of cadmium into quantum dots in Escherichia coli. J Hazard Mater 412:125248
Cai L, Zheng S-W, Shen Y-J, Zheng G-D, Liu H-T, Wu Z-Y (2018) Complete genome sequence provides insights into the biodrying-related microbial function of Bacillus thermoamylovorans isolated from sewage sludge biodrying material. Bioresour Technol 260:141–149
Funding
This work was supported by the National Natural Science Foundation of China (52160015), the Guizhou Provincial Science and Technology Development Project (QKZYD [2022]4022), the Guizhou Hundred Level Talents Project (Guizhou Kehe Platform Talents [2020]6002), the Basic Research Program of Guizhou Province of China (No. ZK[2024]276), and the Guizhou Provincial Science and Technology Projects, (No.[2020]-046).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Yu Zheng: conceptualization, data curation, formal analysis, methodology, software, writing–original draft, and writing–review and editing. Zhibin Duan: conceptualization, supervision, and writing–review and editing. Yonggui Wu: funding acquisition, project administration, resources, and writing–review and editing. Yang Luo: formal analysis, methodology, and writing–review and editing. Xiaoyu Peng: validation and supervision. Jianye Wu: formal analysis and validation. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study focuses on screening and studying bacterial strains, while the specific extraction of bacterial strains from human fecal samples does not require additional ethical approval. As such, it does not involve direct human experimentation or any associated ethical concerns, but we still emphasize our commitment to ethical standards. The human fecal samples used in our study were obtained with proper informed consent procedures and were processed in a way that ensures the complete anonymity of the donors, with no personally identifiable information included. No specific ethical approval was required for this part of the study.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, Y., Duan, Z., Wu, Y. et al. Analysis of the Cadmium Removal Mechanism of Human Gut Bacteria Enterococcus faecalis Strain ATCC19433 from a Genomic Perspective. Biol Trace Elem Res (2024). https://doi.org/10.1007/s12011-024-04169-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12011-024-04169-6