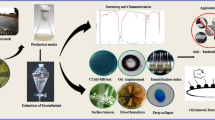
Abstract
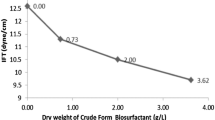
Biosurfactants are renewable resources with versatile applications on environmental bioremediation and industrial processes. Pseudomonas species are one of the promising biosurfactant producers. However, besides rhamnolipids, little is known about Pseudomonas-derived biosurfactants on solubilization of polycyclic aromatic hydrocarbons (PAHs) and oily sludge treatment. In this study, Pseudomonas sp. HN11-derived biosurfactant was purified by chromatographic methods and was characterized as viscosin via bioinformatic analysis, spectrometric and spectroscopic analyses, Marfey’s method and (C-H)α NMR fingerprint matching approach. Viscosin is a potent biosurfactant with critical micelle concentration of 5.79 mg/L and is stable under various stresses. Moreover, viscosin was produced at 0.42 g/L at 48 h of liquid fermentation. Further data have shown that emulsifying agent viscosin is capable of promoting the solubilization of PAHs and displays enhanced oil recovery during oily sludge treatment. More specifically, viscosin has shown significantly enhanced solubilization on fluoranthene compared with control (0.04 mg/L), 2.21 mg/L and 1.27 mg/L fluoranthene was recovered from 100 mg/L and 200 mg/L viscosin treatment, respectively. However, only 200 mg/L viscosin has significantly enhanced the solubilization of phenanthrene (0.75 mg/L) and benzo[a]pyrene (0.51 mg/L) compared to each control (0.23 mg/L for phenanthrene and 0.09 mg/L for benzo[a]pyrene). Viscosin treatment of oily sludge (recovering of 0.58 g oil) has shown a significant oil recovery compared to that of control (recovering of 0.42 g oil). This study shows the great potential of viscosin-type biosurfactant on oily sludge treatment.







Similar content being viewed by others
Data Availability
The genome sequence of Pseudomonas sp. HN11 was deposited in GenBank under accession number CP089985. Pseudomonas sp. HN11 has been deposited in Guangdong Microbial Culture Collection Center (GDMCC), China, with a deposition number GDMCC 1.3638.
References
Carolin, C. F., Kumar, P. S., & Ngueagni, P. T. (2021). A review on new aspects of lipopeptide biosurfactant: Types, production, properties and its application in the bioremediation process. Journal of Hazardous Materials, 407, 124827. https://doi.org/10.1016/j.jhazmat.2020.124827.
Markande, A. R., Patel, D., & Varjani, S. (2021). A review on biosurfactants: Properties, applications and current developments. Bioresource Technology, 330, 124963. https://doi.org/10.1016/j.biortech.2021.124963.
Mishra, S., Lin, Z., Pang, S., Zhang, Y., Bhatt, P., & Chen, S. (2021). Biosurfactant is a powerful tool for the bioremediation of heavy metals from contaminated soils. Journal of Hazardous Materials, 418, 126253. https://doi.org/10.1016/j.jhazmat.2021.126253.
Sun, S., Wang, Y., Zang, T., Wei, J., Wu, H., Wei, C., Qiu, G., & Li, F. (2019). A biosurfactant-producing Pseudomonas aeruginosa S5 isolated from coking wastewater and its application for bioremediation of polycyclic aromatic hydrocarbons. Bioresource Technology, 281, 421–428. https://doi.org/10.1016/j.biortech.2019.02.087.
Zhao, F., Yuan, M., Lei, L., Li, C., & Xu, X. (2021). Enhanced production of mono-rhamnolipid in Pseudomonas aeruginosa and application potential in agriculture and petroleum industry. Bioresource Technology, 323, 124605. https://doi.org/10.1016/j.biortech.2020.124605.
Süssmuth, R. D., & Mainz, A. (2017). Nonribosomal peptide synthesis-principles and prospects. Angewandte Chemie, 56(14), 3770–3821. https://doi.org/10.1002/anie.201609079.
Lopes, L. D., Davis, E. W. 2nd, Pereira, E. S. M. C., Weisberg, A. J., Bresciani, L., Chang, J. H., Loper, J. E., & Andreote, F. D. (2018). Tropical soils are a reservoir for fluorescent Pseudomonas spp. biodiversity. Environmental Microbiology, 20(1), 62–74. https://doi.org/10.1111/1462-2920.13957.
Geudens, N., & Martins, J. C. (2018). Cyclic lipodepsipeptides from Pseudomonas spp. - biological swiss-army knives. Frontiers in Microbiology, 9, 1867. https://doi.org/10.3389/fmicb.2018.01867.
Götze, S., & Stallforth, P. (2020). Structure, properties, and biological functions of nonribosomal lipopeptides from pseudomonads. Natural Product Reports, 37(1), 29–54. https://doi.org/10.1039/c9np00022d.
Geudens, N., De Vleeschouwer, M., Fehér, K., Rokni-Zadeh, H., Ghequire, M. G., Madder, A., De Mot, R., Martins, J. C., & Sinnaeve, D. (2014). Impact of a stereocentre inversion in cyclic lipodepsipeptides from the viscosin group: A comparative study of the viscosinamide and pseudodesmin conformation and self-assembly. Chembiochem : A European Journal Of Chemical Biology, 15(18), 2736–2746. https://doi.org/10.1002/cbic.201402389.
Ma, Z. (2022). Analysis of the complete genome sequence of a rhizosphere-derived Pseudomonas sp. HN3-2 leads to the characterization of a cyclic lipopeptide-type antibiotic bananamide C. 3 Biotech, 12, 35. https://doi.org/10.1007/s13205-021-03100-3
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution, 30(12), 2725–2729. https://doi.org/10.1093/molbev/mst197.
Meier-Kolthoff, J. P., & Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nature Communications, 10(1), 2182. https://doi.org/10.1038/s41467-019-10210-3.
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., & Weber, T. (2021). antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Research, 49(W1), W29–W35. https://doi.org/10.1093/nar/gkab335.
De Roo, V., Verleysen, Y., Kovács, B., De Vleeschouwer, M., Muangkaew, P., Girard, L., Höfte, M., De Mot, R., Madder, A., Geudens, N., & Martins, J. C. (2022). An nuclear magnetic resonance fingerprint matching approach for the identification and structural re-evaluation of Pseudomonas Lipopeptides. Microbiology Spectrum, e0126122. https://doi.org/10.1128/spectrum.01261-22.
Vijayasarathy, S., Prasad, P., Fremlin, L. J., Ratnayake, R., Salim, A. A., Khalil, Z., & Capon, R. J. (2016). C3 and 2D C3 Marfey’s methods for amino acid analysis in natural products. Journal of Natural Products, 79(2), 421–427. https://doi.org/10.1021/acs.jnatprod.5b01125.
Oni, F. E., Geudens, N., Adiobo, A., Omoboye, O. O., Enow, E. A., Onyeka, J. T., Salami, A. E., De Mot, R., Martins, J. C., & Höfte, M. (2020). Biosynthesis and antimicrobial activity of pseudodesmin and viscosinamide cyclic lipopeptides produced by pseudomonads associated with the cocoyam rhizosphere. Microorganisms, 8(7), 1079. https://doi.org/10.3390/microorganisms8071079.
De Bruijn, I., de Kock, M. J. D., Yang, M., de Waard, P., van Beek, T. A., & Raaijmakers, J. M. (2007). Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Molecular Microbiology, 63(2), 417–428. https://doi.org/10.1111/j.1365-2958.2006.05525.x.
Dumonceaux, T. J., Town, J., Links, M. G., & Boyetchko, S. (2014). High-quality draft genome sequence of Pseudomonas sp. BRG100, a strain with bioherbicidal properties against Setaria viridis (Green Foxtail) and other pests of agricultural significance. Genome Announcements, 2(5), e00995–e00914. https://doi.org/10.1128/genomeA.00995-14.
Rokni-Zadeh, H., Li, W., Sanchez-Rodriguez, A., Sinnaeve, D., Rozenski, J., Martins, J. C., & De Mot, R. (2012). Genetic and functional characterization of cyclic lipopeptide white-line-inducing principle (WLIP) production by rice rhizosphere isolate Pseudomonas putida RW10S2. Applied and Environmental Microbiology, 78(14), 4826–4834. https://doi.org/10.1128/AEM.00335-12.
De Bruijn, I., de Kock, M. J. D., de Waard, P., van Beek, T. A., & Raaijmakers, J. M. (2008). Massetolide a biosynthesis in Pseudomonas fluorescens. Journal of Bacteriology, 190(8), 2777–2789. https://doi.org/10.1128/JB.01563-07.
Laycock, M. V., Hildebrand, P. D., Thibault, P., Walter, J. A., & Wright, J. L. C. (1991). Viscosin, a potent peptidolipid biosurfactant and phytopathogenic mediator produced by a pectolytic strain of Pseudomonas fluorescens. Journal of Agricultural and Food Chemistry, 39(3), 483–489. https://doi.org/10.1021/jf00003a011.
Gerard, J., Lloyd, R., Barsby, T., Haden, P., Kelly, M. T., & Andersen, R. J. (1997). Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. Journal of Natural Products, 60(3), 223–229. https://doi.org/10.1021/np9606456.
Oni, F. E., Geudens, N., Omoboye, O. O., Bertier, L., Hua, H. G. K., Adiobo, A., Sinnaeve, D., Martins, J. C., & Höfte, M. (2019). Fluorescent Pseudomonas and cyclic lipopeptide diversity in the rhizosphere of cocoyam (Xanthosoma sagittifolium). Environmental Microbiology, 21(3), 1019–1034. https://doi.org/10.1111/1462-2920.14520.
Oni, F. E., Geudens, N., Onyeka, J. T., Olorunleke, O. F., Salami, A. E., Omoboye, O. O., Arias, A. A., Adiobo, A., De Neve, S., Ongena, M., Martins, J. C., & Höfte, M. (2020b). Cyclic lipopeptide-producing Pseudomonas koreensis group strains dominate the cocoyam rhizosphere of a Pythium root rot suppressive soil contrasting with P. putida prominence in conducive soils. Environmental Microbiology, 22(12), 5137–5155. https://doi.org/10.1111/1462-2920.15127.
Omoboye, O. O., Geudens, N., Duban, M., Chevalier, M., Flahaut, C., Martins, J. C., Leclère, V., Oni, F. E., & Höfte, M. (2019). Pseudomonas sp. COW3 produces new bananamide-type cyclic lipopeptides with antimicrobial activity against Pythium myriotylum and pyricularia oryzae. Molecules, 24(22), 4170. https://doi.org/10.3390/molecules24224170.
Saini, H. S., Barragán-Huerta, B. E., Lebrón-Paler, A., Pemberton, J. E., Vázquez, R. R., Burns, A. M., Marron, M. T., Seliga, C. J., Gunatilaka, A. A., & Maier, R. M. (2008). Efficient purification of the biosurfactant viscosin from Pseudomonas libanensis strain M9-3 and its physicochemical and biological properties. Journal of Natural Products, 71(6), 1011–1015. https://doi.org/10.1021/np800069u.
Gross, H., Stockwell, V. O., Henkels, M. D., Nowak-Thompson, B., Loper, J. E., & Gerwick, W. H. (2007). The genomisotopic approach: A systematic method to isolate products of orphan biosynthetic gene clusters. Chemistry & Biology, 14(1), 53–63. https://doi.org/10.1016/j.chembiol.2006.11.007.
Gerhardt, H., Sievers-Engler, A., Jahanshah, G., Pataj, Z., Ianni, F., Gross, H., Lindner, W., & Lämmerhofer, M. (2016). Methods for the comprehensive structural elucidation of constitution and stereochemistry of lipopeptides. Journal of Chromatography A, 1428, 280–291. https://doi.org/10.1016/j.chroma.2015.05.065.
Ma, Z. (2023). Genome mining and chemical characterization of a new cyclic lipopeptide associated with MDN-0066 from Pseudomonas moraviensis HN2 cultured in a valine-rich medium. The Journal of Antibiotics, in press, https://doi.org/10.1038/s41429-023-00597-z
Liang, X., Guo, C., Liao, C., Liu, S., Wick, L. Y., Peng, D., Yi, X., Lu, G., Yin, H., Lin, Z., & Dang, Z. (2017). Drivers and applications of integrated clean-up technologies for surfactant-enhanced remediation of environments contaminated with polycyclic aromatic hydrocarbons (PAHs). Environmental Pollution, 225, 129–140. https://doi.org/10.1016/j.envpol.2017.03.045.
Kim, C. H., Lee, D. W., Heo, Y. M., Lee, H., Yoo, Y., Kim, G. H., & Kim, J. J. (2019). Desorption and solubilization of anthracene by a rhamnolipid biosurfactant from Rhodococcus fascians. Water Environment Research, 91(8), 739–747. https://doi.org/10.1002/wer.1103.
Li, S., Pi, Y., Bao, M., Zhang, C., Zhao, D., Li, Y., Sun, P., & Lu, J. (2015). Effect of rhamnolipid biosurfactant on solubilization of polycyclic aromatic hydrocarbons. Marine Pollution Bulletin, 101(1), 219–225. https://doi.org/10.1016/j.marpolbul.2015.09.059.
Posada-Baquero, R., Grifoll, M., & Ortega-Calvo, J. J. (2019). Rhamnolipid-enhanced solubilization and biodegradation of PAHs in soils after conventional bioremediation. Science of The Total Environment, 668, 790–796. https://doi.org/10.1016/j.scitotenv.2019.03.056.
Yu, H., Huang, G., Wei, J., & An, C. (2011). Solubilization of mixed polycyclic aromatic hydrocarbons through a rhamnolipid biosurfactant. Journal of Environmental Quality, 40(2), 477–483. https://doi.org/10.2134/jeq2010.0286.
Portet-Koltalo, F., Ammami, M. T., Benamar, A., Wang, H., Derf, F. L., & Duclairoir-Poc, C. (2013). Investigation of the release of PAHs from artificially contaminated sediments using cyclolipopeptidic biosurfactants. Journal of Hazardous Materials, 261, 593–601. https://doi.org/10.1016/j.jhazmat.2013.07.062.
Sanchez-Martin, M. J., Dorado, M. C., & del Hoyo, C.,Rodriguez-Cruz., M.S (2008). Influence of clay mineral structure and surfactant nature on the adsorption capacity of surfactants by clays. Journal of Hazardous Materials, 150(1), 115–123. https://doi.org/10.1016/j.jhazmat.2007.04.093.
Rodriguez-Cruz, M. S., Sanchez-Martin, M. J., & Sanchez-Camazano, M. (2005). A comparative study of adsorption of an anionic and a non-ionic surfactant by soils based on physicochemical and mineralogical properties of soils. Chemosphere, 61(1), 56–64. https://doi.org/10.1016/j.chemosphere.2005.03.016.
Noordman, W. H., Brusseau, M. L., & Janssen, D. B. (2000). Adsorption of a multicomponent rhamnolipid surfactant to soil. Environmental Science and Technology, 34(5), 832–838. https://doi.org/10.1021/es9909982.
Garcia-Junco, M., Gomez-Lahoz, C., Niqui-Arroyo, J. L., & Ortega-Calvo, J. J. (2003). Biosurfactant- and biodegradation-enhanced partitioning of polycyclic aromatic hydrocarbons from nonaqueous-phase liquids. Environmental Science and Technology, 37(13), 2988–2996. https://doi.org/10.1021/es020197q.
Varjani, S., & Upasani, V. N. (2019). Evaluation of rhamnolipid production by a halotolerant novel strain of Pseudomonas aeruginosa. Bioresource Technology, 288, 121577. https://doi.org/10.1016/j.biortech.2019.121577.
Saravanan, A., Kumar, P. S., Vardhan, K. H., Jeevanantham, S., Karishma, S. B., Yaashikaa, P. R., & Vellaichamy, P. (2020). A review on systematic approach for microbial enhanced oil recovery technologies: Opportunities and challenges. Journal of Cleaner Production, 258, 120777. https://doi.org/10.1016/j.jclepro.2020.120777.
Adetunji, A. I., & Olaniran, A. O. (2021). Production and potential biotechnological applications of microbial surfactants: An overview. Saudi Journal of Biological Sciences, 28(1), 669–679. https://doi.org/10.1016/j.sjbs.2020.10.058.
Ke, C. Y., Qin, F. L., Yang, Z. G., Sha, J., Sun, W. J., Hui, J. F., Zhang, Q. Z., & Zhang, X. L. (2021). Bioremediation of oily sludge by solid complex bacterial agent with a combined two-step process. Ecotoxicology and Environmental Safety, 208, 111673. https://doi.org/10.1016/j.ecoenv.2020.111673.
Zheng, C., Wang, M., Wang, Y., & Huang, Z. (2012). Optimization of biosurfactant-mediated oil extraction from oil sludge. Bioresource Technology, 110, 338–342. https://doi.org/10.1016/j.biortech.2012.01.073.
Pacwa-Płociniczak, M., Płaza, G. A., Piotrowska-Seget, Z., & Cameotra, S. S. (2011). Environmental applications of biosurfactants: Recent advances. International Journal of Molecular Sciences, 12(1), 633–654. https://doi.org/10.3390/ijms12010633.
Bao, Q., Huang, L., Xiu, J., Yi, L., & Ma, Y. (2021). Study on the treatment of oily sludge in oil fields with lipopeptide/sophorolipid complex bio-surfactant. Ecotoxicology and Environmental Safety, 212, 111964. https://doi.org/10.1016/j.ecoenv.2021.111964.
Bao, Q., Huang, L., Xiu, J., Yi, L., Zhang, Y., & Wu, B. (2022). Study on the thermal washing of oily sludge used by rhamnolipid/sophorolipid binary mixed bio-surfactant systems. Ecotoxicology and Environmental Safety, 240, 113696. https://doi.org/10.1016/j.ecoenv.2022.113696.
Funding
The first author acknowledges the grants from the National Natural Science Foundation of China (no. 32060625) and the Natural Science Foundation of Gansu Province (no. 20JR5RA527) for supporting this study.
Author information
Authors and Affiliations
Contributions
ZM initiated and designed the study, conducted most experiments (bacterial isolation and identification, genome mining, purification and characterization of biosurfactant, CMC measurements and emulsifying activity tests) and wrote the manuscript. PZ provided the materials (or reagents) for this study. JS, QL and QX were involved in the stability test of the biosurfactant, bacterial fermentation and oily sludge washing experiments. CK provided the materials (or reagents) for this study. All authors have reviewed and approved the final version of the manuscript and authorized the submission of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, Z., Zuo, P., Sheng, J. et al. Characterization and Production of a Biosurfactant Viscosin from Pseudomonas sp. HN11 and its Application on Enhanced oil Recovery During oily Sludge Cleaning. Appl Biochem Biotechnol 195, 7668–7684 (2023). https://doi.org/10.1007/s12010-023-04503-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04503-z