Abstract
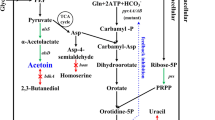
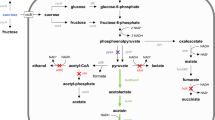
2,3-butanediol is known to be a platform chemical with several potential industrial applications. Sustainable industrial scale production can be attained by using a sugarcane molasses based fermentation process using Bacillus subtilis. However, the accumulation of acetoin needs to be reduced to improve process efficiency. In this work, B. subtilis was genetically modified in order to increase the yield of 2,3-butanediol. Metabolic engineering strategies such as cofactor engineering and overexpression of the key enzyme butanediol dehydrogenase were attempted. Both the strategies individually led to a statistically significant increase in the 2,3-butanediol yields for sugarcane molasses based fermentation. Cofactor engineering led to a 26 % increase in 2,3-butanediol yield and overexpression of bdhA led to a 11 % increase. However, the combination of the two strategies did not lead to a synergistic increase in 2,3-butanediol yield.
Similar content being viewed by others
References
Garg, S. K., & Jain, A. (1995). Fermentative production of 2,3-butanediol: a review. Bioresource Technology, 51, 103–109.
Celińska, E., & Grajek, W. (2009). Biotechnological production of 2,3-butanediol—current state and prospects. Biotechnology Advances, 27, 715–725.
Ji, X. J., Huang, H., & Ouyang, P. K. (2011). Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnology Advances, 29, 351–364.
Ramos, H. C., Hoffmann, T., Marino, M., Nedjari, H., Presecan-Siedel, E., Dreesen, O., Glaser, P., & Jahn, D. (2000). Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression. Journal of Bacteriology, 182, 3072–3080.
Biswas, R., Yamaoka, M., Nakayama, H., Kondo, T., Yoshida, K., Bisaria, V. S., & Kondo, A. (2012). Enhanced production of 2,3-butanediol by engineered Bacillus subtilis. Applied Microbiology and Biotechnology, 94, 651–658.
Deshmukh, A., Mistry, S., Yewale, T., Mahajan, D., & Jain, R. (2015). Production of 2, 3-Butanediol from Sugarcane Molasses Using Bacillus subtilis. International Journal of Advanced Biotechnology and Research, 6, 66–79.
Fu, J., Wang, Z., Chen, T., Liu, W., Shi, T., Wang, G., Tang, Y. J., & Zhao, X. (2014). NADH plays the vital role for chiral pure D-(-)-2,3-butanediol production in Bacillus subtilis under limited oxygen conditions. Biotechnology and Bioengineering, 11, 2126–2131.
Yang T-W, Rao Z-M, Zhang X, Xu M-J, Xu ZH, Yang S-T (2013b). Effects of corn steep liquor on production of 2,3-butanediol and acetoin by Bacillus subtilis. Process Biochemistry, 48, 1610-1617.
Nicholson, W. L. (2008). The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Applied and Environmental Microbiology, 74, 6832–6838.
Wang, Y., San, K. Y., & Bennett, G. N. (2013). Cofactor engineering for advancing chemical biotechnology. Current Opinion in Biotechnology, 24, 994–999.
Yang T, Rao Z, Zhang X, Xu M, Xu Z, Yang ST. (2013a). Improved production of 2,3-butanediol in Bacillus amyloliquefaciens by over-expression of glyceraldehyde-3-phosphate dehydrogenase and 2,3-butanediol dehydrogenase. PLoS One, 8, e76149.
Yang T, Rao Z, Zhang X, Xu M, Xu Z, Yang ST. (2015a). Enhanced 2,3-butanediol production from biodiesel-derived glycerol by engineering of cofactor regeneration and manipulating carbon flux in Bacillus amyloliquefaciens. Microbial Cell Factories, 14, 122-132.
Wang, Y., Li, L., Ma, C., Gao, C., Tao, F., & Xu, P. (2013). Engineering of cofactor regeneration enhances (2S,3S)-2,3-butanediol production from diacetyl. Scientific Reports, 3, 2643–2648.
Donk, W. A., & Zhao, H. (2003). Recent developments in pyridine nucleotide regeneration. Current Opinion in Biotechnology, 14, 421–426.
Gaspar, P., Neves, A. R., Gasson, M. J., Shearman, C. A., & Santos, H. (2011). High yields of 2,3-butanediol and mannitol in Lactococcus lactis through engineering of NAD+ cofactor recycling. Applied and Environmental Microbiology, 77, 6826–6835.
Ehsani, M., Fernández, M. R., Biosca, J. A., Julien, A., & Dequin, S. (2009). Engineering of 2,3-butanediol dehydrogenase to reduce acetoin formation by glycerol-overproducing, low-alcohol Saccharomyces cerevisiae. Applied and Environmental Microbiology, 75, 3196–3205.
Kim, S. J., Seo, S. O., Jin, Y. S., & Seo, J. H. (2013). Production of 2,3-butanediol by engineered Saccharomyces cerevisiae. Bioresource Technology, 146, 274–281.
Vojcic, L., Despotovic, D., Martinez, R., Maurer, K. H., & Schwaneberg, U. (2012). An efficient transformation method for Bacillus subtilis DB104. Applied Microbiology and Biotechnology, 94, 487–493.
Hartig, E., & Jahn, D. (2012). Regulation of the anaerobic metabolism of Bacillus subtilis. Advances in Microbial Physiology, 61, 195–216.
Larsson, J. T., Rogstam, A., & von Wachenfeldt, C. (2005). Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis. Microbiology, 151, 3323–3335.
Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., Bertero, M. G., Bessières, P., Bolotin, A., Borchert, S., Borriss, R., Boursier, L., Brans, A., Braun, M., Brignell, S. C., Bron, S., Brouillet, S., Bruschi, C. V., Caldwell, B., Capuano, V., Carter, N. M., Choi, S. K., Cordani, J. J., Connerton, I. F., Cummings, N. J., Daniel, R. A., Denziot, F., Devine, K. M., Düsterhöft, A., Ehrlich, S. D., Emmerson, P. T., Entian, K. D., Errington, J., Fabret, C., Ferrari, E., Foulger, D., Fritz, C., Fujita, M., Fujita, Y., Fuma, S., Galizzi, A., Galleron, N., Ghim, S. Y., Glaser, P., Goffeau, A., Golightly, E. J., Grandi, G., Guiseppi, G., Guy, B. J., Haga, K., Haiech, J., Harwood, C. R., Hènaut, A., Hilbert, H., Holsappel, S., Hosono, S., Hullo, M. F., Itaya, M., Jones, L., Joris, B., Karamata, D., Kasahara, Y., Klaerr-Blanchard, M., Klein, C., Kobayashi, Y., Koetter, P., Koningstein, G., Krogh, S., Kumano, M., Kurita, K., Lapidus, A., Lardinois, S., Lauber, J., Lazarevic, V., Lee, S. M., Levine, A., Liu, H., Masuda, S., Mauël, C., Médigue, C., Medina, N., Mellado, R. P., Mizuno, M., Moestl, D., Nakai, S., Noback, M., Noone, D., O’Reilly, M., Ogawa, K., Ogiwara, A., Oudega, B., Park, S. H., Parro, V., Pohl, T. M., Portelle, D., Porwollik, S., Prescott, A. M., Presecan, E., Pujic, P., Purnelle, B., Rapoport, G., Rey, M., Reynolds, S., Rieger, M., Rivolta, C., Rocha, E., Roche, B., Rose, M., Sadaie, Y., Sato, T., Scanlan, E., Schleich, S., Schroeter, R., Scoffone, F., Sekiguchi, J., Sekowska, A., Seror, S. J., Serror, P., Shin, B. S., Soldo, B., Sorokin, A., Tacconi, E., Takagi, T., Takahashi, H., Takemaru, K., Takeuchi, M., Tamakoshi, A., Tanaka, T., Terpstra, P., Togoni, A., Tosato, V., Uchiyama, S., Vandebol, M., Vannier, F., Vassarotti, A., Viari, A., Wambutt, R., Wedler, H., Weitzenegger, T., Winters, P., Wipat, A., Yamamoto, H., Yamane, K., Yasumoto, K., Yata, K., Yoshida, K., Yoshikawa, H. F., Zumstein, E., Yoshikawa, H., & Danchin, A. (1997). The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256.
Acknowledgments
The authors thank Dr. Anup Kadam (Analytical sciences department Praj Matrix-R &D center) for co-operation for sample analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Deshmukh, A.N., Nipanikar-Gokhale, P. & Jain, R. Engineering of Bacillus subtilis for the Production of 2,3-Butanediol from Sugarcane Molasses. Appl Biochem Biotechnol 179, 321–331 (2016). https://doi.org/10.1007/s12010-016-1996-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-1996-9