Abstract
Purpose of Review
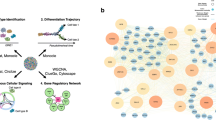
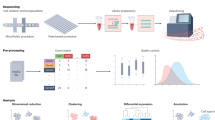
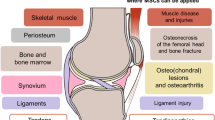
RNA-sequencing (RNA-seq) is a novel and highly sought-after tool in the field of musculoskeletal regenerative medicine. The technology is being used to better understand pathological processes, as well as elucidate mechanisms governing development and regeneration. It has allowed in-depth characterization of stem cell populations and discovery of molecular mechanisms that regulate stem cell development, maintenance, and differentiation in a way that was not possible with previous technology. This review introduces RNA-seq technology and how it has paved the way for advances in musculoskeletal regenerative medicine.
Recent Findings
Recent studies in regenerative medicine have utilized RNA-seq to decipher mechanisms of pathophysiology and identify novel targets for regenerative medicine. The technology has also advanced stem cell biology through in-depth characterization of stem cells, identifying differentiation trajectories and optimizing cell culture conditions. It has also provided new knowledge that has led to improved growth factor use and scaffold design for musculoskeletal regenerative medicine.
Summary
This article reviews recent studies utilizing RNA-seq in the field of musculoskeletal regenerative medicine. It demonstrates how transcriptomic analysis can be used to provide insights that can aid in formulating a regenerative strategy.



Similar content being viewed by others
Abbreviations
- RNA:
-
Ribonucleic acid
- RNA-seq:
-
RNA-sequencing
- mRNAs:
-
Messenger RNAs
- miRNAs:
-
MicroRNAs
- siRNAs:
-
Small interfering RNAs
- NGS:
-
Next-generation sequencing
- cDNA:
-
Complimentary DNA
- scRNA-seq:
-
Single-cell RNA-seq
- GO:
-
Gene ontology
- Obese/T2D:
-
Obese and type 2 diabetes
- MSCs:
-
Mesenchymal stem cells
- ADSC:
-
Adipose-derived stem cells
- BMSC:
-
Bone marrow–derived stem cells
- HLA:
-
Human leukocyte antigen
- WJMSCs:
-
Wharton’s jelly–derived MSCs
- UCMSCs:
-
Umbilical cord MSCs
- ACs:
-
Articular chondrocytes
- PCA:
-
Principal component analysis
- hPSCs:
-
Human pluripotent stem cells
- hiPSCs:
-
Human induced pluripotent stem cells
References
Yelin E, Weinstein S, King T. The burden of musculoskeletal diseases in the United States. Semin Arthritis Rheum. 2016;46(3):259–60.
Vos T, Barber RM, Bell B, Bertozzi-Villa A, Biryukov S, Bolliger I, Charlson F, Davis A, Degenhardt L, Dicker D, Duan L, Erskine H, Feigin VL, Ferrari AJ, Fitzmaurice C, Fleming T, Graetz N, Guinovart C, Haagsma J, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386(9995):743–800.
Ciccocioppo R, Cantore A, Chaimov D, Orlando G. Regenerative medicine: the red planet for clinicians. Intern Emerg Med. 2019;14(6):911–21.
Matichescu A, Ardelean LC, Rusu LC, Craciun D, Bratu EA, Babucea M, Leretter M. Advanced biomaterials and techniques for oral tissue engineering and regeneration—a review. Materials. 2020;13(22):5303.
Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63.
Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82.
Olsen TK, Baryawno N. Introduction to single-cell RNA sequencing. Curr Protoc Mol Biol. 2018;122(1):e57.
Li X, Liao Z, Deng Z, Chen N, Zhao L. Combining bulk and single-cell RNA-sequencing data to reveal gene expression pattern of chondrocytes in the osteoarthritic knee. Bioengineered. 2021;12(1):997–1007.
Huynh NPT, Zhang B, Guilak F. High-depth transcriptomic profiling reveals the temporal gene signature of human mesenchymal stem cells during chondrogenesis. FASEB J. 2019;33(1):358–72.
Doostparast Torshizi A, Wang K. Next-generation sequencing in drug development: target identification and genetically stratified clinical trials. Drug Discov Today. 2018;23(10):1776–83.
Jaksik R, Iwanaszko M, Rzeszowska-Wolny J, Kimmel M. Microarray experiments and factors which affect their reliability. Biol Direct. 2015;10(1):46.
Mehta JP. In: O'Driscoll L, editor. Microarray analysis of mRNAs: experimental design and data analysis fundamentals, in Gene Expression Profiling: Methods and Protocols. Totowa: Humana Press; 2011. p. 27–40.
Bottomly D, Walter NAR, Hunter JE, Darakjian P, Kawane S, Buck KJ, Searles RP, Mooney M, McWeeney SK, Hitzemann R. Evaluating gene expression in C57BL/6J and DBA/2J mouse striatum using RNA-Seq and microarrays. PLoS ONE. 2011;6(3):e17820.
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–17.
Liu Y, Zhou J, White KP. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics. 2014;30(3):301–4.
Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20(11):631–56.
Kukurba KR, Montgomery SB. RNA sequencing and analysis. Cold Spring Harb Protoc. 2015;2015(11) p. pdb.top084970.
Auer PL, Doerge RW. Statistical design and analysis of RNA sequencing data. Genetics. 2010;185(2):405–16.
Fang Z, Cui X. Design and validation issues in RNA-seq experiments. Brief Bioinform. 2011;12(3):280–7.
Koch CM, Chiu SF, Akbarpour M, Bharat A, Ridge KM, Bartom ET, Winter DR. A beginner’s guide to analysis of RNA sequencing data. Am J Respir Cell Mol Biol. 2018;59(2):145–57.
Yalamanchili HK, Wan YW, Liu Z. Data analysis pipeline for RNA-seq experiments: from differential expression to cryptic splicing. Curr Protoc Bioinform. 2017;59(1):11.15.1–11.15.21.
Munger SC, Raghupathy N, Choi K, Simons AK, Gatti DM, Hinerfeld DA, Svenson KL, Keller MP, Attie AD, Hibbs MA, Graber JH, Chesler EJ, Churchill GA. RNA-Seq alignment to individualized genomes improves transcript abundance estimates in multiparent populations. Genetics. 2014;198(1):59–73.
Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21(12):2213–23.
Raghupathy N, Choi K, Vincent MJ, Beane GL, Sheppard KS, Munger SC, Korstanje R, Pardo-Manual de Villena F, Churchill GA. Hierarchical analysis of RNA-seq reads improves the accuracy of allele-specific expression. Bioinformatics. 2018;34(13):2177–84.
Oikonomopoulos S, Bayega A, Fahiminiya S, Djambazian H, Berube P, Ragoussis J. Methodologies for transcript profiling using long-read technologies. Front Genet. 2020;11:606–6.
Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502.
Choi YH, Kim JK. Dissecting cellular heterogeneity using single-cell RNA sequencing. Mol Cell. 2019;42(3):189–99.
Sun C, Wang L, Wang H, Huang T, Yao W, Li J, Zhang X. Single-cell RNA-seq highlights heterogeneity in human primary Wharton’s jelly mesenchymal stem/stromal cells cultured in vitro. Stem Cell Res Ther. 2020;11(1):149.
Zhong L, Yao L, Tower RJ, Wei Y, Miao Z, Park J, Shrestha R, Wang L, Yu W, Holdreith N, Huang X, Zhang Y, Tong W, Gong Y, Ahn J, Susztak K, Dyment N, Li M, Long F, et al. Single cell transcriptomics identifies a unique adipose lineage cell population that regulates bone marrow environment. eLife. 2020;9.
Peng G, Cui G, Ke J, Jing N. Using single-cell and spatial transcriptomes to understand stem cell lineage specification during early embryo development. Annu Rev Genomics Hum Genet. 2020;21(1):163–81.
Brielle S, Bavli D, Motzik A, Kan-Tor Y, Sun X, Kozulin C, Avni B, Ram O, Buxboim A. Delineating the heterogeneity of matrix-directed differentiation toward soft and stiff tissue lineages via single-cell profiling. Proc Natl Acad Sci. 2021;118(19):e2016322118.
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–20.
Lafzi A, Moutinho C, Picelli S, Heyn H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat Protoc. 2018;13(12):2742–57.
Lee JS, Yi K, Ju YS, Shin EC. Effects of Cryopreservation and Thawing on Single-Cell Transcriptomes of Human T Cells. Immune Netw. 2020;20(4):e34.
Denisenko E, Guo BB, Jones M, Hou R, de Kock L, Lassmann T, Poppe D, Clément O, Simmons RK, Lister R, Forrest ARR. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020;21(1):130.
Andrews TS, Kiselev VY, McCarthy D, Hemberg M. Tutorial: guidelines for the computational analysis of single-cell RNA sequencing data. Nat Protoc. 2021;16(1):1–9.
Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29.
Tuerlings M, et al. RNA sequencing reveals interacting key determinants of osteoarthritis acting in subchondral bone and articular cartilage: identification of IL11 and CHADL as attractive treatment targets. Arthritis Rheum. 2021;73:789–99.
Coutinho De Almeida R, et al. RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage. Ann Rheum Dis. 2019;78(2):270–7.
Ji Q, Zheng Y, Zhang G, Hu Y, Fan X, Hou Y, Wen L, Li L, Xu Y, Wang Y, Tang F. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann Rheum Dis. 2019;78(1):100–10.
Dell'Orso S, et al. Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development. 2019;146(12) p. dev174177.
Qu F, Palte IC, Gontarz PM, Zhang B, Guilak F. Transcriptomic analysis of bone and fibrous tissue morphogenesis during digit tip regeneration in the adult mouse. FASEB J. 2020;34(7):9740–54.
Qin T, Fan CM, Wang TZ, Sun H, Zhao YY, Yan RJ, Yang L, Shen WL, Lin JX, Bunpetch V, Cucchiarini M, Clement ND, Mason CE, Nakamura N, Bhonde R, Yin Z, Chen X. Single-cell RNA-seq reveals novel mitochondria-related musculoskeletal cell populations during adult axolotl limb regeneration process. Cell Death Differ. 2021;28(3):1110–25.
Song N, Scholtemeijer M, Shah K. Mesenchymal stem cell immunomodulation: mechanisms and therapeutic potential. Trends Pharmacol Sci. 2020;41(9):653–64.
Pino CJ, Westover AJ, Johnston KA, Buffington DA, Humes HD. Regenerative medicine and immunomodulatory therapy: insights from the kidney, heart, brain, and lung. Kidney Int Rep. 2018;3(4):771–83.
Guerrouahen BS, Sidahmed H, al Sulaiti A, al Khulaifi M, Cugno C. Enhancing mesenchymal stromal cell immunomodulation for treating conditions influenced by the immune system. Stem Cells Int. 2019;2019:1–11.
Lee DK, Song SU. Immunomodulatory mechanisms of mesenchymal stem cells and their therapeutic applications. Cell Immunol. 2018;326:68–76.
Panina Y, Karagiannis P, Kurtz A, Stacey GN, Fujibuchi W. Human Cell Atlas and cell-type authentication for regenerative medicine. Exp Mol Med. 2020;52(9):1443–51.
Andrews TS, Hemberg M. Identifying cell populations with scRNASeq. Mol Asp Med. 2018;59:114–22.
Savulescu AF, Jacobs C, Negishi Y, Davignon L, Mhlanga MM. Pinpointing cell identity in time and space. Front Mol Biosci. 2020;7.
Zhou W, Lin J, Zhao K, Jin K, He Q, Hu Y, Feng G, Cai Y, Xia C, Liu H, Shen W, Hu X, Ouyang H. Single-cell profiles and clinically useful properties of human mesenchymal stem cells of adipose and bone marrow origin. Am J Sports Med. 2019;47(7):1722–33.
Sun C, Zhang K, Yue J, Meng S, Zhang X. Deconstructing transcriptional variations and their effects on immunomodulatory function among human mesenchymal stromal cells. Stem Cell Res Ther. 2021;12(1):53.
Stüdle C, Occhetta P, Geier F, Mehrkens A, Barbero A, Martin I. Challenges toward the identification of predictive markers for human mesenchymal stromal cells chondrogenic potential. Stem Cells Transl Med. 2019;8(2):194–204.
Yin Z, Lin J, Yan R, Liu R, Liu M, Zhou B, Zhou W, An C, Chen Y, Hu Y, Fan C, Zhao K, Wu B, Zou X, Zhang J, el-Hashash AH, Chen X, Ouyang H. Atlas of musculoskeletal stem cells with the soft and hard tissue differentiation architecture. Advanced Science. 2020;7(23):2000938.
Baldwin MJ, Cribbs AP, Guilak F, Snelling SJB. Mapping the musculoskeletal system one cell at a time. Nat Rev Rheumatol. 2021;17(5):247–8.
Wu C-L, Dicks A, Steward N, Tang R, Katz DB, Choi YR, Guilak F. Single cell transcriptomic analysis of human pluripotent stem cell chondrogenesis. Nat Commun. 2021;12(1):362.
Kelly NH, Huynh NPT, Guilak F. Single cell RNA-sequencing reveals cellular heterogeneity and trajectories of lineage specification during murine embryonic limb development. Matrix Biol. 2020;89:1–10.
Wolock SL, et al. Mapping distinct bone marrow niche populations and their differentiation paths. Cell Rep. 2019;28(2):302–311.e5.
Han X, Chen H, Huang D, Chen H, Fei L, Cheng C, Huang H, Yuan GC, Guo G. Mapping human pluripotent stem cell differentiation pathways using high throughput single-cell RNA-sequencing. Genome Biol. 2018;19(1):47.
Elabd C, Ichim TE, Miller K, Anneling A, Grinstein V, Vargas V, Silva FJ. Comparing atmospheric and hypoxic cultured mesenchymal stem cell transcriptome: implication for stem cell therapies targeting intervertebral discs. J Transl Med. 2018;16(1):222.
Shaik S, Martin EC, Hayes DJ, Gimble JM, Devireddy RV. Transcriptomic profiling of adipose derived stem cells undergoing osteogenesis by RNA-Seq. Sci Rep. 2019;9(1):11800.
Chang C-C, Venø MT, Chen L, Ditzel N, le DQS, Dillschneider P, Kassem M, Kjems J. Global MicroRNA profiling in human bone marrow skeletal—stromal or mesenchymal–stem cells identified candidates for bone regeneration. Mol Ther. 2018;26(2):593–605.
Carrow JK, Cross LM, Reese RW, Jaiswal MK, Gregory CA, Kaunas R, Singh I, Gaharwar AK. Widespread changes in transcriptome profile of human mesenchymal stem cells induced by two-dimensional nanosilicates. Proc Natl Acad Sci. 2018;115(17):E3905–13.
Bao X, Shi R, Zhao T, Wang Y, Anastasov N, Rosemann M, Fang W. Integrated analysis of single-cell RNA-seq and bulk RNA-seq unravels tumour heterogeneity plus M2-like tumour-associated macrophage infiltration and aggressiveness in TNBC. Cancer Immunol Immunother. 2021;70(1):189–202.
Method of the Year 2020: spatially resolved transcriptomics. Nat Methods. 2021;18(1):1.
Larsson L, Frisén J, Lundeberg J. Spatially resolved transcriptomics adds a new dimension to genomics. Nat Methods. 2021;18(1):15–8.
Merritt CR, Ong GT, Church SE, Barker K, Danaher P, Geiss G, Hoang M, Jung J, Liang Y, McKay-Fleisch J, Nguyen K, Norgaard Z, Sorg K, Sprague I, Warren C, Warren S, Webster PJ, Zhou Z, Zollinger DR, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. 2020;38(5):586–99.
Sikes KJ, McConnell A, Serkova N, Cole B, Frisbie D. Untargeted metabolomics analysis identifies creatine, myo-inositol, and lipid pathway modulation in a murine model of tendinopathy. J Orthop Res. 2021.
Schott EM, Farnsworth CW, Grier A, Lillis JA, Soniwala S, Dadourian GH, Bell RD, Doolittle ML, Villani DA, Awad H, Ketz JP, Kamal F, Ackert-Bicknell C, Ashton JM, Gill SR, Mooney RA, Zuscik MJ. Targeting the gut microbiome to treat the osteoarthritis of obesity. JCI Insight. 2018;3(8):e95997.
Funding
This work was supported by the National Institutes of Health under award numbers R33HD090696 (KAP), R01AR079839 (CLAB), R01AR078414 (MJZ), and the Department of Defense under award number W81XWH-19-1-0807 (MJZ)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Regenerative Biology and Medicine in Osteoporosis
Rights and permissions
About this article
Cite this article
Thomas, S.M., Ackert-Bicknell, C.L., Zuscik, M.J. et al. Understanding the Transcriptomic Landscape to Drive New Innovations in Musculoskeletal Regenerative Medicine. Curr Osteoporos Rep 20, 141–152 (2022). https://doi.org/10.1007/s11914-022-00726-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11914-022-00726-x