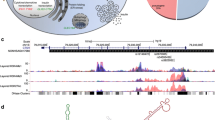
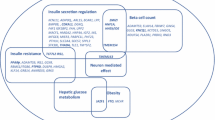
Abstract
Purpose of Review
To provide an updated summary of discoveries made to date resulting from genome-wide association study (GWAS) and sequencing studies, and to discuss the latest loci added to the growing repertoire of genetic signals predisposing to type 1 diabetes (T1D).
Recent Findings
Genetic studies have identified over 60 loci associated with T1D susceptibility. GWAS alone does not specifically inform on underlying mechanisms, but in combination with other sequencing and omics-data, advances are being made in our understanding of T1D genetic etiology and pathogenesis. Current knowledge indicates that genetic variation operating in both pancreatic β cells and in immune cells is central in mediating T1D risk.
Summary
One of the main challenges is to determine how these recently discovered GWAS-implicated variants affect the expression and function of gene products. Once we understand the mechanism of action for disease-causing variants, we will be well placed to apply targeted genomic approaches to impede the premature activation of the immune system in an effort to ultimately prevent the onset of T1D.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Steyn NP, Lambert EV, Tabana H. Conference on “multidisciplinary approaches to nutritional problems”. Symposium on “diabetes and health”. Nutrition interventions for the prevention of type 2 diabetes. Proc Nutr Soc. 2009;68(1):55–70. https://doi.org/10.1017/S0029665108008823.
EURODIAB ACE Study Group. Variation and trends in incidence of childhood diabetes in Europe. Lancet. 2000;355(9207):873–6.
Onkamo P, Vaananen S, Karvonen M, Tuomilehto J. Worldwide increase in incidence of type I diabetes–the analysis of the data on published incidence trends. Diabetologia. 1999;42(12):1395–403. https://doi.org/10.1007/s001250051309.
You WP, Henneberg M. Type 1 diabetes prevalence increasing globally and regionally: the role of natural selection and life expectancy at birth. BMJ Open Diabetes Res Care. 2016;4(1):e000161. https://doi.org/10.1136/bmjdrc-2015-000161.
Redondo MJ, Yu L, Hawa M, Mackenzie T, Pyke DA, Eisenbarth GS, et al. Heterogeneity of type I diabetes: analysis of monozygotic twins in Great Britain and the United States. Diabetologia. 2001;44(3):354–62. https://doi.org/10.1007/s001250051626.
Hyttinen V, Kaprio J, Kinnunen L, Koskenvuo M, Tuomilehto J. Genetic liability of type 1 diabetes and the onset age among 22,650 young Finnish twin pairs: a nationwide follow-up study. Diabetes. 2003;52(4):1052–5.
Olmos P, A'Hern R, Heaton DA, Millward BA, Risley D, Pyke DA, et al. The significance of the concordance rate for type 1 (insulin-dependent) diabetes in identical twins. Diabetologia. 1988;31(10):747–50.
Kyvik KO, Green A, Beck-Nielsen H. Concordance rates of insulin dependent diabetes mellitus: a population based study of young Danish twins. BMJ. 1995;311(7010):913–7.
Clayton DG. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genet. 2009;5(7):e1000540. https://doi.org/10.1371/journal.pgen.1000540.
Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet. 2016;387(10035):2340–8. https://doi.org/10.1016/S0140-6736(16)30507-4.
Boljat A, Gunjaca I, Konstantinovic I, Vidan N, Boraska Perica V, Pehlic M, et al. Environmental risk factors for type 1 diabetes mellitus development. Exp Clin Endocrinol Diabetes. 2017;125(8):563–70. https://doi.org/10.1055/s-0043-109000.
Paul DS, Teschendorff AE, Dang MA, Lowe R, Hawa MI, Ecker S, et al. Increased DNA methylation variability in type 1 diabetes across three immune effector cell types. Nat Commun. 2016;7:13555. https://doi.org/10.1038/ncomms13555.
Cudworth AG, Woodrow JC. Evidence for HL-A-linked genes in “juvenile” diabetes mellitus. Br Med J. 1975;3(5976):133–5.
Nerup J, Platz P, Andersen OO, Christy M, Lyngsoe J, Poulsen JE, et al. HL-A antigens and diabetes mellitus. Lancet. 1974;2(7885):864–6.
Singal DP, Blajchman MA. Histocompatibility (HL-A) antigens, lymphocytotoxic antibodies and tissue antibodies in patients with diabetes mellitus. Diabetes. 1973;22(6):429–32.
Melanitou E, Fain P, Eisenbarth GS. Genetics of type 1A (immune mediated) diabetes. J Autoimmun. 2003;21(2):93–8.
Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57(4):1084–92. https://doi.org/10.2337/db07-1331.
Bell GI, Horita S, Karam JH. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes. 1984;33(2):176–83.
Nistico L, Buzzetti R, Pritchard LE, Van der Auwera B, Giovannini C, Bosi E, et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Belgian Diabetes Registry. Hum Mol Genet. 1996;5(7):1075–80.
Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36(4):337–8. https://doi.org/10.1038/ng1323.
Vella A, Cooper JD, Lowe CE, Walker N, Nutland S, Widmer B, et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76(5):773–9. https://doi.org/10.1086/429843.
Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38(6):617–9. https://doi.org/10.1038/ng1800.
Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448(7153):591–4. https://doi.org/10.1038/nature06010.
Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. https://doi.org/10.1038/nature05911.
Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet. 1996;59(5):1134–48.
Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39(7):857–64. https://doi.org/10.1038/ng2068.
Concannon P, Onengut-Gumuscu S, Todd JA, Smyth DJ, Pociot F, Bergholdt R, et al. A human type 1 diabetes susceptibility locus maps to chromosome 21q22.3. Diabetes. 2008;57(10):2858–61. https://doi.org/10.2337/db08-0753.
Manolio TA, Rodriguez LL, Brooks L, Abecasis G, Ballinger D, Daly M, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39(9):1045–51. https://doi.org/10.1038/ng2127.
Mueller PW, Rogus JJ, Cleary PA, Zhao Y, Smiles AM, Steffes MW, et al. Genetics of Kidneys in Diabetes (GoKinD) study: a genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J Am Soc Nephrol. 2006;17(7):1782–90. https://doi.org/10.1681/ASN.2005080822.
Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40(12):1399–401. https://doi.org/10.1038/ng.249.
Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–7. https://doi.org/10.1038/ng.381.
Cooper JD, Walker NM, Healy BC, Smyth DJ, Downes K, Todd JA. Analysis of 55 autoimmune disease and type II diabetes loci: further confirmation of chromosomes 4q27, 12q13.2 and 12q24.13 as type I diabetes loci, and support for a new locus, 12q13.3-q14.1. Genes Immun. 2009;10(Suppl 1):S95–120. https://doi.org/10.1038/gene.2009.98.
Fung EY, Smyth DJ, Howson JM, Cooper JD, Walker NM, Stevens H, et al. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun. 2009;10(2):188–91. https://doi.org/10.1038/gene.2008.99.
Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359(26):2767–77. https://doi.org/10.1056/NEJMoa0807917.
Bakay M, Pandey R, Hakonarson H. Genes involved in type 1 diabetes: an update. Genes (Basel). 2013;4(3):499–521. https://doi.org/10.3390/genes4030499.
Qu HQ, Bradfield JP, Li Q, Kim C, Frackelton E, Grant SF, et al. In silico replication of the genome-wide association results of the Type 1 Diabetes Genetics Consortium. Hum Mol Genet. 2010;19(12):2534–8. https://doi.org/10.1093/hmg/ddq133.
Wang K, Baldassano R, Zhang H, Qu HQ, Imielinski M, Kugathasan S, et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum Mol Genet. 2010;19(10):2059–67. https://doi.org/10.1093/hmg/ddq078.
Cooper JD, Howson JM, Smyth D, Walker NM, Stevens H, Yang JH, et al. Confirmation of novel type 1 diabetes risk loci in families. Diabetologia. 2012;55(4):996–1000. https://doi.org/10.1007/s00125-012-2450-3.
Bradfield JP, Qu HQ, Wang K, Zhang H, Sleiman PM, Kim CE, et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet. 2011;7(9):e1002293. https://doi.org/10.1371/journal.pgen.1002293.
Hakonarson H, Qu HQ, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, et al. A novel susceptibility locus for type 1 diabetes on chr12q13 identified by a genome-wide association study. Diabetes 2008;(57):1143–1146
Grant SF, Qu HQ, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Taback SP, Frackelton, EC Eckert AW, et al. Follow-up analysis of genome-wide association data identifies nvel loci for type1 diabetes. Diabetes 2009;(58):209–295
Awata T, Kawasaki E, Tanaka S, Ikegami H, Maruyama T, Shimada A, Nakanishi K, Kobayashi T, Lizuka H, Uga M, et al. Association of type 1 diabetes with two loci on 12q13 and 16p13 and the inluence coexisting thyroid autoimmunity in japanese. j Clin Endocrinol Metab 2009;(94):231–235
Zoledziewska M, Costa G, Pitzalis M, Cocco E, Melis C, Moi L, et al. Variation within the CLEC16A gene shows consistent disease association with both multiple sclerosis and type 1 diabetes in Sardinia. Genes Immun. 2009;10(1):15–7. https://doi.org/10.1038/gene.2008.84.
Wu X, Zhu X, Wang X, Ma J, Zhu S, Li J, et al. Intron polymorphism in the KIAA0350 gene is reproducibly associated with susceptibility to type 1 diabetes (T1D) in the Han Chinese population. Clin Endocrinol. 2009;71(1):46–9. https://doi.org/10.1111/j.1365-2265.2008.03437.x.
Wallace C, Smyth DJ, Maisuria-Armer M, walker NM, Todd JA, clayton DG. The imrinted dlk1-meg3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes Nat Genet 2010;(42):68–71
Reddy MV, Wang H, Liu S, Bode B, Reed JC, Steed RD, et al. Association between type 1 diabetes and GWAS SNPs in the southeast US Caucasian population. Genes Immun. 2011;12(3):208–12. https://doi.org/10.1038/gene.2010.70.
Asad S, Nikamo P, Gyllenberg A, Bennet H, Hansson O, wierup N, Carlsson A, Forsander G, Ivarsoon SA, Larsonn H, et al. Htr1a a novel type 1 diabetes susceptibility gene on chromosome 5p13-q13. Plos One 2012;(7):e35439
Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1,000 genomes-based implutation identifies novel and refines associations for the wellcome trust case control consortium phase 1 data. Eur J Hum Genet 2012;(20):801–805
• Sharma A, Liu X, Hadley D, Hagopian W, Chen WM, Onengut-Gumuscu S, et al. Identification of non-HLA genes associated with development of islet autoimmunity and type 1 diabetes in the prospective TEDDY cohort. J Autoimmun. 2018;89:90–100. https://doi.org/10.1016/j.jaut.2017.12.008. This study uses large progressive cohorts to identify novel genetic factors for islet autoimmunity and T1D.
Ahlqvist E, van Zuydam NR, Groop LC, McCarthy MI. The genetics of diabetic complications. Nat Rev Nephrol. 2015;11(5):277–87. https://doi.org/10.1038/nrneph.2015.37.
Craig DW, Millis MP, DiStefano JK. Genome-wide SNP genotyping study using pooled DNA to identify candidate markers mediating susceptibility to end-stage renal disease attributed to type 1 diabetes. Diabet Med. 2009;26(11):1090–8. https://doi.org/10.1111/j.1464-5491.2009.02846.x.
Williams WW, Salem RM, McKnight AJ, Sandholm N, Forsblom C, Taylor A, et al. Association testing of previously reported variants in a large case-control meta-analysis of diabetic nephropathy. Diabetes. 2012;61(8):2187–94. https://doi.org/10.2337/db11-0751.
Sandholm N, Salem RM, McKnight AJ, Brennan EP, Forsblom C, Isakova T, et al. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 2012;8(9):e1002921. https://doi.org/10.1371/journal.pgen.1002921.
Sandholm N, McKnight AJ, Salem RM, Brennan EP, Forsblom C, Harjutsalo V, et al. Chromosome 2q31.1 associates with ESRD in women with type 1 diabetes. J Am Soc Nephrol. 2013;24(10):1537–43. https://doi.org/10.1681/ASN.2012111122.
Hotaling JM, Waggott DR, Goldberg J, Jarvik G, Paterson AD, Cleary PA, et al. Pilot genome-wide association search identifies potential loci for risk of erectile dysfunction in type 1 diabetes using the DCCT/EDIC study cohort. J Urol. 2012;188(2):514–20. https://doi.org/10.1016/j.juro.2012.04.001.
Meng W, Veluchamy A, Hebert HL, Campbell A, Colhoun HM, Palmer CNA. A genome-wide association study suggests that MAPK14 is associated with diabetic foot ulcers. Br J Dermatol. 2017;177(6):1664–70. https://doi.org/10.1111/bjd.15787.
Charmet R, Duffy S, Keshavarzi S, Gyorgy B, Marre M, Rossing P, et al. Novel risk genes identified in a genome-wide association study for coronary artery disease in patients with type 1 diabetes. Cardiovasc Diabetol. 2018;17(1):61. https://doi.org/10.1186/s12933-018-0705-0.
Wallace C, Cutler AJ, Pontikos N, Pekalski ML, Burren OS, Cooper JD, et al. Dissection of a complex disease susceptibility region using a Bayesian stochastic search approach to fine mapping. PLoS Genet. 2015;11(6):e1005272. https://doi.org/10.1371/journal.pgen.1005272.
Onengut-Gumuscu S, Chen WM, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47(4):381–6. https://doi.org/10.1038/ng.3245.
Cotsapas C, Hafler DA. Immune-mediated disease genetics: the shared basis of pathogenesis. Trends Immunol. 2013;34(1):22–6. https://doi.org/10.1016/j.it.2012.09.001.
Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7(8):e1002254. https://doi.org/10.1371/journal.pgen.1002254.
Li YR, Li J, Zhao SD, Bradfield JP, Mentch FD, Maggadottir SM, et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat Med. 2015;21(9):1018–27. https://doi.org/10.1038/nm.3933.
Tomer Y, Dolan LM, Kahaly G, Divers J, D'Agostino RB Jr, Imperatore G, et al. Genome wide identification of new genes and pathways in patients with both autoimmune thyroiditis and type 1 diabetes. J Autoimmun. 2015;60:32–9. https://doi.org/10.1016/j.jaut.2015.03.006.
Westra HJ, Martinez-Bonet M, Onengut-Gumuscu S, Lee A, Luo Y, Teslovich N, et al. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat Genet. 2018;50(10):1366–74. https://doi.org/10.1038/s41588-018-0216-7.
Ram R, Mehta M, Nguyen QT, Larma I, Boehm BO, Pociot F, et al. Systematic evaluation of genes and genetic variants associated with type 1 diabetes susceptibility. J Immunol. 2016;196(7):3043–53. https://doi.org/10.4049/jimmunol.1502056.
Pociot F, Kaur S, Nielsen LB. Effects of the genome on immune regulation in type 1 diabetes. Pediatr Diabetes. 2016;17(Suppl 22):37–42. https://doi.org/10.1111/pedi.12336.
Eizirik DL, Sammeth M, Bouckenooghe T, Bottu G, Sisino G, Igoillo-Esteve M, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8(3):e1002552. https://doi.org/10.1371/journal.pgen.1002552.
Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41(2):827–41. https://doi.org/10.1093/nar/gks1284.
Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22(9):1748–59. https://doi.org/10.1101/gr.136127.111.
Cerosaletti K, Barahmand-Pour-Whitman F, Yang J, DeBerg HA, Dufort MJ, Murray SA, et al. Single-cell RNA sequencing reveals expanded clones of islet antigen-reactive CD4(+) T cells in peripheral blood of subjects with type 1 diabetes. J Immunol. 2017;199(1):323–35. https://doi.org/10.4049/jimmunol.1700172.
Leung CS, Yang KY, Li X, Chan VW, Ku M, Waldmann H, et al. Single-cell transcriptomics reveal that PD-1 mediates immune tolerance by regulating proliferation of regulatory T cells. Genome Med. 2018;10(1):71. https://doi.org/10.1186/s13073-018-0581-y.
Mastracci TL, Turatsinze JV, Book BK, Restrepo IA, Pugia MJ, Wiebke EA, et al. Distinct gene expression pathways in islets from individuals with short- and long-duration type 1 diabetes. Diabetes Obes Metab. 2018;20(8):1859–67. https://doi.org/10.1111/dom.13298.
•• Ahmed R, Omidian Z, Giwa A, Cornwell B, Majety N, Bell DR, et al. A public BCR present in a unique dual-receptor-expressing lymphocyte from type 1 diabetes patients encodes a potent T cell autoantigen. Cell. 2019;177(6):1583–99 e16. https://doi.org/10.1016/j.cell.2019.05.007. This study employed scRNA-seq to investigate blood of T1D and control individuals. It discovered a rare subset of lymphocytes that coexpresses T and B cell markers and expands in T1D.
Lu JM, Chen YC, Ao ZX, Shen J, Zeng CP, Lin X, et al. System network analysis of genomics and transcriptomics data identified type 1 diabetes-associated pathway and genes. Genes Immun. 2018;20:500–8. https://doi.org/10.1038/s41435-018-0045-9.
Nyaga DM, Vickers MH, Jefferies C, Perry JK, O'Sullivan JM. Type 1 diabetes mellitus-associated genetic variants contribute to overlapping immune regulatory networks. Front Genet. 2018;9:535. https://doi.org/10.3389/fgene.2018.00535.
Gao P, Uzun Y, He B, Salamati SE, Coffey JKM, Tsalikian E, et al. Risk variants disrupting enhancers of TH1 and TREG cells in type 1 diabetes. Proc Natl Acad Sci U S A. 2019;116(15):7581–90. https://doi.org/10.1073/pnas.1815336116.
Sharma A, Liu X, Hadley D, Hagopian W, Liu E, Chen WM, et al. Identification of non-HLA genes associated with celiac disease and country-specific differences in a large, international pediatric cohort. PLoS One. 2016;11(3):e0152476. https://doi.org/10.1371/journal.pone.0152476.
Cervin C, Lyssenko V, Bakhtadze E, Lindholm E, Nilsson P, Tuomi T, et al. Genetic similarities between latent autoimmune diabetes in adults, type 1 diabetes, and type 2 diabetes. Diabetes. 2008;57(5):1433–7. https://doi.org/10.2337/db07-0299.
• Cousminer DL, Ahlqvist E, Mishra R, Andersen MK, Chesi A, Hawa MI, et al. First genome-wide association study of latent autoimmune diabetes in adults reveals novel insights linking immune and metabolic diabetes. Diabetes Care. 2018;41(11):2396–403. https://doi.org/10.2337/dc18-1032. The first GWAS of LADA. This study confirmed that the strongest genetic risk loci for LADA are common with T1D, while also showed positive genetic correlations with T2D.
Mishra R, Chesi A, Cousminer DL, Hawa MI, Bradfield JP, Hodge KM, et al. Relative contribution of type 1 and type 2 diabetes loci to the genetic etiology of adult-onset, non-insulin-requiring autoimmune diabetes. BMC Med. 2017;15(1):88. https://doi.org/10.1186/s12916-017-0846-0.
Thomas NJ, Jones SE, Weedon MN, Shields BM, Oram RA, Hattersley AT. Frequency and phenotype of type 1 diabetes in the first six decades of life: a cross-sectional, genetically stratified survival analysis from UK Biobank. Lancet Diabetes Endocrinol. 2018;6(2):122–9. https://doi.org/10.1016/S2213-8587(17)30362-5.
Redondo MJ, Steck AK, Sosenko J, Anderson M, Antinozzi P, Michels A, et al. Transcription factor 7-like 2 (TCF7L2) gene polymorphism and progression from single to multiple autoantibody positivity in individuals at risk for type 1 diabetes. Diabetes Care. 2018;41(12):2480–6. https://doi.org/10.2337/dc18-0861.
Redondo MJ, Geyer S, Steck AK, Sosenko J, Anderson M, Antinozzi P, et al. TCF7L2 genetic variants contribute to phenotypic heterogeneity of type 1 diabetes. Diabetes Care. 2018;41(2):311–7. https://doi.org/10.2337/dc17-0961.
Martinez A, Perdigones N, Cenit MC, Espino L, Varade J, Lamas JR, et al. Chromosomal region 16p13: further evidence of increased predisposition to immune diseases. Ann Rheum Dis. 2010;69(1):309–11. https://doi.org/10.1136/ard.2008.098376.
Sang Y, Zong W, Yan J, Liu M. The correlation between the CLEC16A gene and genetic susceptibility to type 1 diabetes in Chinese children. Int J Endocrinol. 2012;2012:245384. https://doi.org/10.1155/2012/245384.
Yamashita H, Awata T, Kawasaki E, Ikegami H, Tanaka S, Maruyama T, et al. Analysis of the HLA and non-HLA susceptibility loci in Japanese type 1 diabetes. Diabetes Metab Res Rev. 2011;27(8):844–8. https://doi.org/10.1002/dmrr.1234.
Howson JM, Rosinger S, Smyth DJ, Boehm BO, Todd JA. Genetic analysis of adult-onset autoimmune diabetes. Diabetes. 2011;60(10):2645–53. https://doi.org/10.2337/db11-0364.
Nischwitz S, Cepok S, Kroner A, Wolf C, Knop M, Muller-Sarnowski F, et al. More CLEC16A gene variants associated with multiple sclerosis. Acta Neurol Scand. 2011;123(6):400–6. https://doi.org/10.1111/j.1600-0404.2010.01421.x.
Zuvich RL, Bush WS, McCauley JL, Beecham AH, De Jager PL, Ivinson AJ, et al. Interrogating the complex role of chromosome 16p13.13 in multiple sclerosis susceptibility: independent genetic signals in the CIITA-CLEC16A-SOCS1 gene complex. Hum Mol Genet. 2011;20(17):3517–24. https://doi.org/10.1093/hmg/ddr250.
Skinningsrud B, Husebye ES, Pearce SH, McDonald DO, Brandal K, Wolff AB, et al. Polymorphisms in CLEC16A and CIITA at 16p13 are associated with primary adrenal insufficiency. J Clin Endocrinol Metab. 2008;93(9):3310–7. https://doi.org/10.1210/jc.2008-0821.
Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1228–33. https://doi.org/10.1038/ng.468.
Zhang Z, Cheng Y, Zhou X, Li Y, Gao J, Han J, et al. Polymorphisms at 16p13 are associated with systemic lupus erythematosus in the Chinese population. J Med Genet. 2011;48(1):69–72. https://doi.org/10.1136/jmg.2010.077859.
Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42(4):295–302. https://doi.org/10.1038/ng.543.
Marquez A, Varade J, Robledo G, Martinez A, Mendoza JL, Taxonera C, et al. Specific association of a CLEC16A/KIAA0350 polymorphism with NOD2/CARD15(−) Crohn’s disease patients. Eur J Hum Genet. 2009;17(10):1304–8. https://doi.org/10.1038/ejhg.2009.50.
Jagielska D, Redler S, Brockschmidt FF, Herold C, Pasternack SM, Garcia Bartels N, et al. Follow-up study of the first genome-wide association scan in alopecia areata: IL13 and KIAA0350 as susceptibility loci supported with genome-wide significance. J Investig Dermatol. 2012;132(9):2192–7. https://doi.org/10.1038/jid.2012.129.
Skinningsrud B, Lie BA, Husebye ES, Kvien TK, Forre O, Flato B, et al. A CLEC16A variant confers risk for juvenile idiopathic arthritis and anti-cyclic citrullinated peptide antibody negative rheumatoid arthritis. Ann Rheum Dis. 2010;69(8):1471–4. https://doi.org/10.1136/ard.2009.114934.
Hirschfield GM, Xie G, Lu E, Sun Y, Juran BD, Chellappa V, et al. Association of primary biliary cirrhosis with variants in the CLEC16A, SOCS1, SPIB and SIAE immunomodulatory genes. Genes Immun. 2012;13(4):328–35. https://doi.org/10.1038/gene.2011.89.
Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43(4):329–32. https://doi.org/10.1038/ng.789.
Berge T, Leikfoss IS, Harbo HF. From identification to characterization of the multiple sclerosis susceptibility gene CLEC16A. Int J Mol Sci. 2013;14(3):4476–97. https://doi.org/10.3390/ijms14034476.
Chang CH, Guerder S, Hong SC, van Ewijk W, Flavell RA. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 1996;4(2):167–78.
Leikfoss IS, Mero IL, Dahle MK, Lie BA, Harbo HF, Spurkland A, et al. Multiple sclerosis-associated single-nucleotide polymorphisms in CLEC16A correlate with reduced SOCS1 and DEXI expression in the thymus. Genes Immun. 2013;14(1):62–6. https://doi.org/10.1038/gene.2012.52.
Leikfoss IS, Keshari PK, Gustavsen MW, Bjolgerud A, Brorson IS, Celius EG, et al. Multiple sclerosis risk allele in CLEC16A acts as an expression quantitative trait locus for CLEC16A and SOCS1 in CD4+ T cells. PLoS One. 2015;10(7):e0132957. https://doi.org/10.1371/journal.pone.0132957.
Davison LJ, Wallace C, Cooper JD, Cope NF, Wilson NK, Smyth DJ, et al. Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene. Hum Mol Genet. 2012;21(2):322–33. https://doi.org/10.1093/hmg/ddr468.
Kim S, Naylor SA, DiAntonio A. Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function. Proc Natl Acad Sci U S A. 2012;109(18):E1072–81. https://doi.org/10.1073/pnas.1120320109.
Soleimanpour SA, Gupta A, Bakay M, Ferrari AM, Groff DN, Fadista J, et al. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell. 2014;157(7):1577–90. https://doi.org/10.1016/j.cell.2014.05.016.
Pearson G, Chai B, Vozheiko T, Liu X, Kandarpa M, Piper RC, et al. Clec16a, Nrdp1, and USP8 form a ubiquitin-dependent tripartite complex that regulates beta-cell mitophagy. Diabetes. 2018;67(2):265–77. https://doi.org/10.2337/db17-0321.
van Luijn MM, Kreft KL, Jongsma ML, Mes SW, Wierenga-Wolf AF, van Meurs M, et al. Multiple sclerosis-associated CLEC16A controls HLA class II expression via late endosome biogenesis. Brain. 2015;138(Pt 6):1531–47. https://doi.org/10.1093/brain/awv080.
Schuster C, Gerold KD, Schober K, Probst L, Boerner K, Kim MJ, et al. The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity. 2015;42(5):942–52. https://doi.org/10.1016/j.immuni.2015.04.011.
Pandey R, Bakay M, Hain HS, Strenkowski B, Elsaqa BZB, Roizen JD, et al. CLEC16A regulates splenocyte and NK cell function in part through MEK signaling. PLoS One. 2018;13(9):e0203952. https://doi.org/10.1371/journal.pone.0203952.
Pandey R, Bakay M, Hain HS, Strenkowski B, Yermakova A, Kushner JA, et al. The autoimmune disorder susceptibility gene CLEC16A restrains NK cell function in YTS NK cell line and Clec16a knockout mice. Front Immunol. 2019;10:68. https://doi.org/10.3389/fimmu.2019.00068.
Redmann V, Lamb CA, Hwang S, Orchard RC, Kim S, Razi M, et al. Clec16a is critical for autolysosome function and Purkinje cell survival. Sci Rep. 2016;6:23326. https://doi.org/10.1038/srep23326.
Harris BS, Ward-Bailey, P. F., Bergstrom, D. E., Bronson, R. T. & Donahue, L. R. Curvy tail: a new skeletal mutation that maps to chromosome 16. Available at: http://www.informatics.jax.org/reference/J:172931. 2011.
Bian X, Wasserfall C, Wallstrom G, Wang J, Wang H, Barker K, et al. Tracking the antibody immunome in type 1 diabetes using protein arrays. J Proteome Res. 2017;16(1):195–203. https://doi.org/10.1021/acs.jproteome.6b00354.
Hu Z, Liu Y, Zhai X, Dai J, Jin G, Wang L, et al. New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet. 2013;45(12):1499–503. https://doi.org/10.1038/ng.2809.
Lee SM, Yang H, Tartar DM, Gao B, Luo X, Ye SQ, et al. Prevention and treatment of diabetes with resveratrol in a non-obese mouse model of type 1 diabetes. Diabetologia. 2011;54(5):1136–46. https://doi.org/10.1007/s00125-011-2064-1.
Agardh E, Lundstig A, Perfilyev A, Volkov P, Freiburghaus T, Lindholm E, et al. Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy. BMC Med. 2015;13:182. https://doi.org/10.1186/s12916-015-0421-5.
Dodge R, Loomans C, Sharma A, Bonner-Weir S. Developmental pathways during in vitro progression of human islet neogenesis. Differentiation. 2009;77(2):135–47. https://doi.org/10.1016/j.diff.2008.09.015.
Funding
This work was supported by an Institute Development Award to the Center for Applied Genomics from the Children’s Hospital of Philadelphia and by the Endowed Chair in Genomic Research to Dr. Hakonarson. Dr. Grant is funded by the Daniel B. Burke Endowed Chair for Diabetes Research, and by NIH grant R01 DK085212.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Marina Bakay and Rahul Pandey report they have a patent pending on new innovative weight reduction therapies targeting CLEC16A.
Struan F.A. grant reports he has a patent issue on genetic alterations and methods of use thereof for the diagnosis and treatment of type I diabetes issued (patent number 10125395); a patent issued on genetic alterations on chromosome 16 and methods of use thereof for the diagnosis and treatment of type 1 diabetes (patent number 10266896); and a patent issued on genetic alterations on chromosomes 21Q, 6Q, and 15Q and methods of use thereof for the diagnosis and treatment of type 1 diabetes (patent number 10066266).
Hakon Hakonarson reports he has a patent issued on genetic alterations and methods of use thereof for the diagnosis and treatment of type I diabetes issued (patent number 10125395); a patent issued on genetic alterations on chromosome 16 and methods of use thereof for the diagnosis and treatment of type 1 diabetes issued (patent number 10266896); a patent issued on genetic alterations on chromosomes 21Q, 6Q, and 15Q and methods of use thereof for the diagnosis and treatment of type 1 diabetes (patent number 10066266); and a patent pending on new innovative weight reduction therapies targeting CLEC16A.
Human and Animal Rights and Informed Consent
All reported studies/experiments with human or animal subjects performed by the authors have been previously published and complied with all applicable ethical standards (including the Helsinki declaration and its amendments, institutional/national research committee standards, and international/national/institutional guidelines).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Genetics
Rights and permissions
About this article
Cite this article
Bakay, M., Pandey, R., Grant, S.F. et al. The Genetic Contribution to Type 1 Diabetes. Curr Diab Rep 19, 116 (2019). https://doi.org/10.1007/s11892-019-1235-1
Published:
DOI: https://doi.org/10.1007/s11892-019-1235-1