Abstract
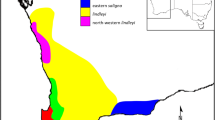
As a dynamic ex situ conservation strategy, a clonal seed orchard was started in a nursery in Pomaio (POM) in central Italy in 1993 for an assisted migration experiment of Abies nebrodensis (Lojac.) Mattei. Two artificial ex situ populations were planted with this gene pool: a seedling arboretum in Pieve Santo Stefano (PSS) and a small dendrological collection in Papiano (PAP), both originating from the Sicilian relict population. Here, using AFLP markers, we estimated the relatedness among the relocated genotypes of the three collections to check whether the three collections had sufficient genetic variability to be considered as additional sources of variability to the original gene pool for the assisted migration strategy. High individual genetic variability was found in the collections; each plant had a different genotype and was confirmed to belong to its population of origin. PAP and PSS trees were shown to be only from the original population of A. nebrodensis species and were derived from a limited set of maternal fertile genotypes. Based on the Sicilian fir population inventory, nursery production in Sicily, and structure clustering analysis, close genetic relationships among POM, PAP and several PSS trees (1–35) were evident. Similarly, the PSS group (36–78) was genetically close to tree 1 of POM and in a lesser proportion to plants 7 and 9 of POM. The sampling of seedlings used to form batches in the nursery might have influenced the structure of the resultant plantations. All genotypes will be useful for enriching the original gene pool.



Similar content being viewed by others
References
Acheré V, Favre JM, Besnard G, Jeandroz S (2005) Genomic organization of molecular differentiation in Norway spruce (Picea abies). Mol Ecol 14:3191–3201
Ahn JY, Lee JW, Lee MW, Hong KN (2019) Genetic diversity and structure of Carpinus laxiflora populations in South Korea based on AFLP markers. Forest Sci Technol 15(4):192–201
Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA (2007) Standardizing methods to address clonality in population Studies. Mol Ecol 16:5115–5139
Aussenac G (2002) Ecology and ecophysiology of circum-Mediterranean firs in the context of climate change. Ann for Sci 59:823–832
Biondi E, Raimondo FM (1980) Primo rinvenimento di legni fossili sulle Madonie. Giorn Bot Ital 114(3–4):128–129
Bobo-Pinilla J, Salmerón-Sánchez E, Mendoza-Fernández AJ, Mota JF, Peñas J (2022) Conservation and phylogeography of plants: from the Mediterranean to the rest of the world. Diversity 14(78):1–23
Bonin A, Ehrich D, Mantel S (2007) Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Mol Ecol 16:3737–3758
Bou Dagher-Kharrat M, Mariette S, François L, Fady B, Grenier-de March G, Plomion CH, Savouré A (2007) Geographical diversity and genetic relationships among Cedrus species estimated by AFLP. Tree Genet Genomes 3:275–285
Cervera MT, Remington D, Frigerio JM, Storme V, Ivens B, Boerjan W, Plomion C (2000) Improved AFLP analysis of tree species. Can J for Res 30(10):1608–1616
Costa P, Pot D, Dubos C, Frigerio JM, Pionneau C, Bodenes C, Bertocchi E, Cervera MT, Remington DL, Plomion C (2000) A genetic map of Maritime pine based on AFLP, RAPD and protein markers. Theor Appl Genet 100:39–48
Costa R, Pereira G, Garrido I, Tavares-de-Sousa MM, Espinosa F (2016) Comparison of RAPD, ISSR, and AFLP molecular markers to reveal and classify orchardgrass (Dactylis glomerata L.) germplasm variations. PLoS ONE 11(4):1–15
Dent AE, Von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4: 359–361
Di XY, Li XN, Wang QX, Wang MB (2014) Genetic diversity of natural populations of Larix principis-rupprechtii in Shanxi Province China. Biochem Syst Ecol 54:71–77
Di XY, Wang MB (2013) Genetic diversity and structure of natural Pinus tabulaeformis populations in North China using amplified fragment length polymorphism (AFLP). Biochem Syst Ecol 51:269–275
Ducci F (2014) Species restoration approach, chapter 151 Species restoration through dynamic ex situ conservation: Abies nebrodensis as a model. In: Genetic considerations in ecosystem restoration using native tree species State of the World’s Forest Genetic Resources – Thematic Study Ed Bozzano et al Rome FAO and Bioversity International, pp 225–232
Ducci F, Proietti R, Favre JM (1999) Allozyme assessment of genetic diversity within the relic Sicilian fir Abies nebrodensis (Lojac.) Mattei. Ann for Sci 56:345–355
Ehrich D, Gaudeul M, Assefa A, ehrich Koch MA, Mummenhoff K, Nemomissa S, Intrabiodiv Consortium, Brochmann C, 2007 Ehrich D, Gaudeul M, Assefa A,ehrich Koch MA, Mummenhoff K, Nemomissa S, Intrabiodiv Consortium, Brochmann C (2007) Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Mol Ecol 16(12): 2542–2559
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14 (8): 2611–2620
Fady B, Aravanopoulos FA, Alizoti P, Mátyás C, von Wühlisch G, Westergren M, Belletti P, Cvjetkovic B, Ducci F, Huber G, Kelleher CT, Khaldi A, Dagher Kharrat MB, Kraigher H, Kramer K, Mühlethaler U, Peric S, Perry A, Rousi M, Sbay H, Stojnic S, Tijardovic M, Tsvetkov I, Varela MC, Vendramin GG, Zlatanov T (2016) Evolution-based approach needed for the conservation and silviculture of peripheral forest tree populations. Forest Ecol Manag 375: 66–75
Farjon A (2018) The Kew review: Conifers of the world. Kew Bull 73(8).
Felsenstein J (2004) Inferring phylogenies. Book Sinauer Associates Inc Publishers Sunderland Massachusetts
Fuentes-Utrilla P, Valbuena-Carabana M, Ennos R, Gil L (2014) Population clustering and clonal structure evidence the relict state of Ulmus minor Mill. in the Balearic Islands. Heredity 113: 21–31
Gaudeul M, Rouhan G, Gardner MF, Hollingsworth PM (2012) AFLP markers provide insights into the evolutionary relationships and diversification of New Caledonian Araucaria species (Araucariaceae). Am J of Bot 99(1):68–81
Gerber S, Mariette S, Streiff R, Bodénès C, Kremer A (2000) Comparison of microsatellites and amplified fragment length polymorphism markers for parentage analysis. Mol Ecol 9:1037–1048
Gussone G (1843) Florae Siculae synopsis. I-Il. - Napoli P II: pp. 647; add. pp. 883
Hardy OJ (2003) Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Mol Ecol 12:1577–1588
Holland BR, Clarke AC, Meudt HM (2008) Optimizing automated AFLP scoring parameters to improve phylogenetic resolution. Syst Biol 57(3):347–366
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9 (5):1322–1332
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23 (14): 1801–1806
Jump AS, Hunt JM, Martinez-Izquierdo JA, Peñuelas J (2006) Natural selection and climate change: temperature-linked spatial and temporal trends in gene frequency in Fagus sylvatica. Mol Ecol 15:3469–3480
Jump AS, Peñuelas J (2007) Extensive spatial genetic structure revealed by AFLP but not SSR molecular markers in the wind pollinated tree, Fagus sylvatica. Mol Ecol 16:925–936
Kim MS, Richardson BA, McDonald GI, Klopfenstein NB (2011) Genetic diversity and structure of western white pine (Pinus monticola) in North America: a baseline study for conservation, restoration, and addressing impacts of climate change. Tree Genet Genomes 7:11–21
Kopelman NM, Mayzel J, Jakobsson M , Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15(5):1179–91
Krauss SL (2000) Accurate gene diversity estimates from amplified fragment length polymorphism (AFLP) markers. Mol Ecol 9:1241–1245
Latch EK, Dharmarajan G, Glaubitz JC, Olin ER, Rhodes JR (2006) Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet 7:295–302
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Mariette S, Chagneâ D, Leâzier C, Pastuska P, Raffin A, Plomion C, Kremer A (2001) Genetic diversity within and among Pinus pinaster populations: comparison between AFLP and microsatellite markers. Heredity 86:469–479
Meirmans PG, Van Tienderen PH (2004) GenoType and GenoDive: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Méndez-González ID, Jardón-Barbolla L, Jaramillo-Correa JP (2017) Differential landscape effects on the fine-scale genetic structure of populations of a montane Conifer from central Mexico. Tree Genet Genomes 13:30
Meudt HM, Clarke AC (2007) Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci12 (3)
Morandini R (1969) Abies nebrodensis (Lojac) Mattei: Inventario 1968. Pubb Istit Sper Selvic 18:1–93
Morandini R, Ducci F, Menguzzato G (1994) Abies nebrodensis (Lojac) Mattei–Inventario 1992. Ann Ist Sper Selv XXII:5–51
Paglia G, Morgante M (1998) PCR-based multiplex DNA fingerprinting technique for the analysis of Conifer genome. Mol Breed 4:173–177
Parducci L, Szmidt AE, Madaghiele A, Anzidei M, Vendramin GG (2001) Genetic variation at chloroplast microsatellites (cpSSRs) in Abies nebrodensis (Lojac) Mattei and three neighboring Abies species. Theor Appl Genet 102:733–740
Pasta S, Sala G, La Mantia T, Bondì C, Tinner W (2020) The past distribution of Abies nebrodensis (Lojac) Mattei: results of a multidisciplinary study. Veget Hist Archaeobot 29:357–371
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pluess AR, Weber P (2012) Drought-adaptation potential in Fagus sylvatica: linking moisture availability with genetic diversity and dendrochronology. PLoS ONE 7(3):1–8
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2010–2012) Documentation for STRUCTURE software Available with the program at: http://pritchbsduchicagoedu/structurehtm
Reisch C, Bernhardt-Römermann M (2014) The impact of study design and life history traits on genetic variation of plants determined with AFLPs. Plant Ecol 215(12):1493–1511
Ribeiro MM, Mariette S, Vendramin GG, Szmidt AE, Plomion C, Kremer A (2002) Comparison of genetic diversity estimates within and among populations of maritime pine using chloroplast simple-sequence repeat and amplified fragment length polymorphism data. Mol Ecol 11:869–877
Ritland K (2005) Multilocus estimation of pairwise relatedness with dominant markers. Mol Ecol 14:3157–3165
Roncallo PF, Beaufort V, Larsen AO, Dreisigacker S, Echenique V (2019) Genetic diversity and linkage disequilibrium using SNP (KASP) and AFLP markers in a worldwide durum wheat (Triticum turgidum L var durum) collection. PLoS ONE 14(6)
Sánchez-Gómez P, Jiménez JF, Cánovas1 JL, Vera1 JB, Hensen I, Aouissat M (2018) Genetic structure and phylogeography of Juniperus phoenicea complex throughout Mediterranean and Macaronesian regions: different stories in one. Ann For Sci 75:75
Sánchez-Robles JM, Balao F, Terrab A, Garcia-Castano JL, Ortiz MA, Vela E, Talavera S (2014) Phylogeography of SW Mediterranean firs: different European origins for the North African Abies species. Mol Phylogenet Evol 79:42–53
Schicchi R, Bazan G, Raimondo F (1997) La progenie di Abies nebrodensis (Lojac) Mattei accertata in vivaio. Quad Bot Ambient Appl 8:3–9
Schlüter PhM, Harris SA (2006) Analysis of multilocus fingerprinting data sets containing missing data. Mol Ecol Notes 6:569–572
Schwartz MK, McKelvey KS (2009) Why sampling scheme matters: the effect of sampling scheme on landscape genetic results. Conserv Genet 10:441–452
Scotti I, González-Martínez SC, Budde KB, Lalagüe H (2016) Fifty years of genetic studies: What to make of the large amounts of variation found within populations? Ann for Sci 73:69–75
Semerikova SA, Lascoux M, Semerikov VL (2012) Nuclear and cytoplasmic genetic diversity reveals long-term population decline in Abies semenovii, an endemic fir of central Asia. Can J For Res 42:2142–2152
Semerikova SA, Semerikov VL (2011) Genetic variability of Siberian fir Abies sibirica Ledeb inferred from AFLP markers. Russ J Genet 47 (2):241–246
Semerikova SA, Semerikov VL (2016) Phylogeny of firs (genus Abies, Pinaceae) based on multilocus nuclear markers (AFLP). Russ J Genet 52 (11):1164–1175
Semerikova SA, Semerikov VL, Lascoux M (2011) Post-glacial history and introgression in Abies (Pinaceae) species of the Russian Far East inferred from both nuclear and cytoplasmic markers. J Biogeogr 38:326–340
Sokal RR, Rohlf FJ (1995) Biometry: the principles and practice of statistics in biological research 3rd Edition WH Freeman and Co New York: pp 896
Tang SQ, Dai WJ, Li MS, Zhang Y, Geng YP, Wang L, Zhong Y (2008) Genetic diversity of relictual and endangered plant Abies ziyuanensis (Pinaceae) revealed by AFLP and SSR markers. Genetica 133:21–30
Thomas P (2017) Abies nebrodensis. The IUCN red list of threatened species 2017: e.T30478A91164876. Downloaded on 10 November 2021. https://doi.org/10.2305/IUCN.UK.2017-2.RLTS.T30478A91164876.en
Vanden Broeck A, Cox K, Melosik I, Maes B, Smets K (2018) Genetic diversity loss and homogenization in urban trees: the case of Tilia × europaea in Belgium and the Netherlands. Biodivers Conserv 27:3777–3792
Vekemans X, Beauwens T, Roldçn-Ruiz I LM (2002) Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11:139–151
Venturella G, Mazzola P, Raimondo FM (1997) Strategies for the conservation and restoration of the relict population of Abies nebrodensis (Lojac.) Mattei Bocconea 7:417–425
Virgilio F, Schicchi R, La Mela Veca DS (2000) Aggiornamento dell’inventario della popolazione relitta di Abies nebrodensis (Lojac) Mattei. Naturalista Sicil S IV XXIV 1–2:13–54
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acid Res 23:4407–4414
Wang J, Abbott RJ, Ingvarsson PK, Liu JQ (2014) Increased genetic divergence between two closely related fir species in areas of range overlap. Ecol Evol 4 (7):1019–1029
Wang XR, Chhatre VE, Nillson MC, Song W, Zackrisson O, Szmidt AE (2003) Island population structure of Norway Spruce (Picea abies) in northern Sweden. Int J Plant Sci 164 (5):711–717
Wessa P (2021) Free statistics software office for research development and education V 1.2.1 URL: https://wwwwessanet/
Williams MI, Dumroese RK (2013) Preparing for climate change: forestry and assisted migration. J For 111: 287–297
Woodhead W, Russell J, Squirrell J, Hollingsworth PM, Mackenzie K, M Gibby M, Powell W (2005) Comparative analysis of population genetic structure in Athyrium distentifolium (Pteridophyta) using AFLPs and SSRs from anonymous and transcribed gene regions. Mol Ecol 14:1681–1695
Xu SQ, Tauer CG, Nelson CD (2008) Genetic diversity within and among populations of shortleaf pine (Pinus echinata Mill.) and loblolly pine (Pinus taeda L.). Tree Genet Genomes 4(4):859–868
Xue XM, Wang YH, Korpelainen H, Li CY (2005) Assessment of AFLP-based genetic variation in the populations of Picea asperata. Silvae Genet 54 (1):24–30
Yang AH, Wei N, Fritsch PW, Yao XH (2016) AFLP genome scanning reveals divergent selection in natural populations of Liriodendron chinense (Magnoliaceae) along a latitudinal transect front. Plant Sci 7:698
Zelener N, Marcucci Poltri SN, Bartoloni N, López CR, Hopp HE (2005) Selection strategy for a seedling seed orchard design based on trait selection index and genomic analysis by molecular markers: a case study for Eucalyptus dunnii. Tree Physiol 25:1457–1467
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Acknowledgements
We thank Dr Juan Fernandez and Prof Nathalie Frascaria-Lacoste for suggestions and help developing the AFLP analysis protocol, and Dr Simona Pecchioli and Dr Silvia Carnevale for doing the molecular analyses at CREA FL Lab.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: Our research was funded by the Italian Ministry for Agriculture, Food and Forestry Policies in the framework of the “FAO-RGV (FAO-Vegetal Genetic Resources) Project”.
The online version is available at http://www.springerlink.com.
Corresponding editor: Tao Xu.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
De Rogatis, A., Ducci, F., Guerri, S. et al. Genotyping ex situ trees of Abies nebrodensis translocated from the original Sicilian population to enrich the gene pool. J. For. Res. 34, 1095–1106 (2023). https://doi.org/10.1007/s11676-022-01534-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-022-01534-w