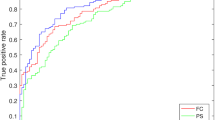
Abstract
Attention deficit/hyperactivity disorder (ADHD) is a common disorder among children. ADHD often prevails into adulthood, unless proper treatments are facilitated to engage self-regulatory systems. Thus, there is a need for effective and reliable mechanisms for the early identification of ADHD. This paper presents a decision support system for the ADHD identification process. The proposed system uses both functional magnetic resonance imaging (fMRI) data and eye movement data. The classification processes contain enhanced pipelines, and consist of pre-processing, feature extraction, and feature selection mechanisms. fMRI data are processed by extracting seed-based correlation features in default mode network (DMN) and eye movement data using aggregated features of fixations and saccades. For the classification using eye movement data, an ensemble model is obtained with 81% overall accuracy. For the fMRI classification, a convolutional neural network (CNN) is used with 82% accuracy for the ADHD identification. Both ensemble models are proved for overfitting avoidance.
Similar content being viewed by others
References
C. Sridhar, S. Bhat, U. R. Acharya, H. Adeli, G. M. Bairy. Diagnosis of attention deficit hyperactivity disorder using imaging and signal processing techniques. Computers in Biology and Medicine, vol. 88, pp. 93–99, 2017. DOI: https://doi.org/10.1016/j.compbiomed.2017.07.009.
D. A. Meedeniya, I. D. Rubasinghe. A review of supportive computational approaches for neurological disorder identification. Interdisciplinary Approaches to Altering Neurodevelopmental Disorders, T. Wadhera, D. Kakkar, Eds., IGI global, Chapter 16, Hershey, USA: IGI Global, pp. 271–302, 2020. DOI: https://doi.org/10.4018/978-1-7998-3069-6.ch016.
B. Zablotsky, L. I. Black, M. J. Maenner, L. A. Schieve, M. L. Danielson, R. H. Bitsko, S. J. Blumberg, M. D. Kogan, C. A. Boyle. Prevalence and trends of developmental disabilities among children in the United States: 2009–2017. Pediatrics, vol. 144, no. 4, Article number e20190811, 2019. DOI: https://doi.org/10.1542/peds.2019-0811.
S. De Silva, S. Dayarathna, G. Ariyarathne, D. Meedeniya, S. Jayarathna. A survey of attention deficit hyperactivity disorder identification using psychophysiological data. International Journal of Online and Biomedical Engineering, vol. 15, no. 13, pp. 61–76, 2019. DOI: https://doi.org/10.3991/ijoe.v15i13.10744.
L. Q. Uddin, A. M. C. Kelly, B. B. Biswal, D. S. Margulies, Z. Shehzad, D. Shaw, M. Ghaffari, J. Rotrosen, L. A. Adler, F. X. Castellanos, M. P. Milham. Network homogeneity reveals decreased integrity of default-mode network in ADHD. Journal of Neuroscience Methods, vol. 169, no. 1, pp. 249–254, 2008. DOI: https://doi.org/10.1016/j.jneumeth.2007.11.031.
I. D. Rubasinghe, D. A. Meedeniya. Automated neuroscience decision support framework. Deep Learning Techniques for Biomedical and Health Informatics, B. Agarwal, V. E. Balas, L. C. Jain, R. C. Poonia, Manisha, Eds., Cambridge, USA: Academic Press, pp.305–326, 2020. DOI: https://doi.org/10.1016/b978-0-12-819061-6.00013-6.
A. M. P. Michalek, G. Jayawardena, S. Jayarathna. Predicting ADHD using eye gaze metrics indexing working memory capacity. Computational Models for Biomedical Reasoning and Problem Solving, C. H. Chen, S. C. S. Cheung, Eds., Hershey: IGI Global, Chapter 3, pp. 66–88, 2019. DOI: https://doi.org/10.4018/978-1-5225-7467-5.ch003.
N. N. J. Rommelse, S. Van Der Stigchel, J. A. Sergeant. A review on eye movement studies in childhood and adolescent psychiatry. Brain and Cognition, vol. 68, no. 3, pp. 391–414, 2008. DOI: https://doi.org/10.1016/j.bandc.2008.08.025.
P. Deans, L. O’Laughlin, B. Brubaker, N. Gay, D. Krug. Use of eye movement tracking in the differential diagnosis of attention deficit hyperactivity disorder (ADHD) and reading disability. Psychology, vol. 1, no. 4, pp. 238–246, 2010. DOI: https://doi.org/10.4236/psych.2010.14032.
S. Van Der Stigchel, M. Meeter, J. Theeuwes. Eye movement trajectories and what they tell us. Neuroscience & Biobehavioral Reviews, vol. 30, no. 5, pp. 666–679, 2006. DOI: https://doi.org/10.1016/j.neubiorev.2005.12.001.
S. De Silva, S. Dayarathna, G. Ariyarathne, D. Meedeniya, S. Jayarathna, A. M. P. Michalek, G. Jayawardena. A rule-based system for ADHD identification using eye movement data. In Proceedings of Moratuwa Engineering Research Conference, IEEE, Moratuwa, Sri Lanka, pp. 538–543, 2019. DOI: https://doi.org/10.1109/mercon.2019.8818865.
K. Krejtz, A. T. Duchowski, A. Niedzielska, C. Biele, I. Krejtz. Eye tracking cognitive load using pupil diameter and microsaccades with fixed gaze. PLoS One, vol. 13, no. 9, Article number e0203629, 2018. DOI: https://doi.org/10.1371/journal.pone.0203629.
M. Fried, E. Tsitsiashvili, Y. S. Bonneh, A. Sterkin, T. Wygnanski-Jaffe, T. Epstein, U. Polat. ADHD subjects fail to suppress eye blinks and microsaccades while anticipating visual stimuli but recover with medication. Vision Research, vol. 101, pp. 62–72, 2014. DOI: https://doi.org/10.1016/j.visres.2014.05.004.
R. G. Ross, J. G. Harris, A. Olincy, A. Radant. Eye movement task measures inhibition and spatial working memory in adults with schizophrenia, ADHD, and a normal comparison group. Psychiatry Research, vol. 95, no. 1, pp. 35–42, 2000. DOI: https://doi.org/10.1016/s0165-1781(00)00153-0.
R. G. Ross, A. Olincy, J. G. Harris, B. Sullivan, A. Radant. Smooth pursuit eye movements in schizophrenia and attentional dysfunction: Adults with schizophrenia, ADHD, and a normal comparison group. Biological Psychiatry, vol. 48, no. 3, pp. 197–203, 2000. DOI: https://doi.org/10.1016/s0006-3223(00)00825-8.
D. P. Munoz, I. T. Armstrong, K. A. Hampton, K. D. Moore. Altered control of visual fixation and saccadic eye movements in attention-deficit hyperactivity disorder. Journal of Neurophysiology, vol. 90, no. 1, pp. 503–514, 2003. DOI: https://doi.org/10.1152/jn.00192.2003.
G. J. Hyun, J. W. Park, J. H. Kim, K. J. Min, Y. S. Lee, S. M. Kim, D. H. Han. Visuospatial working memory assessment using a digital tablet in adolescents with attention deficit hyperactivity disorder. Computer Methods and Programs in Biomedicine, vol. 157, pp. 137–143, 2018. DOI: https://doi.org/10.1016/j.cmpb.2018.01.022.
I. H. Witten, E. Frank, M. A. Hall, C. J. Pal. Data Mining: Practical Machine Learning Tools and Techniques, 4th ed., San Francisco, USA: Morgan Kaufmann, 2016.
M. Kantardzic. Data Mining: Concepts, Models, Methods, and Algorithms, 3rd ed., Hoboken, USA: John Wiley & Sons, 2019.
Y. H. Shi, W. M. Zeng, N. Z. Wang, D. T. L. Chen. A novel fMRI group data analysis method based on data-driven reference extracting from group subjects. Computer Methods and Programs in Biomedicine, vol. 122, no. 3, pp. 362–371, 2015. DOI: https://doi.org/10.1016/j.cmpb.2015.09.002.
NITRC: ADHD-200, 2019, [Online], Available: https://www.nitrc.org/ir/app/template/, May 2, 2020.
B. Jie, M. X. Liu, D. G. Shen. Integration of temporal and spatial properties of dynamic connectivity networks for automatic diagnosis of brain disease. Medical Image Analysis, vol. 47, pp. 81–94, 2018. DOI: https://doi.org/10.1016/j.media.2018.03.013.
S. De Silva, S. Dayarathna, G. Ariyarathne, D. Meedeniya, S. Jayarathna. fMRI feature extraction model for ADHD classification using convolutional neural network. International Journal of E-Health and Medical Communications, vol. 12, no. 1, pp. 81–105, 2021. DOI: https://doi.org/10.4018/IJEHMC.2021010106.
L. Zou, J. N. Zheng, C. Y. Miao, M. Mckeown, Z. J. Wang. 3D CNN based automatic diagnosis of attention deficit hyperactivity disorder using functional and structural MRI. IEEE Access, vol. 5, pp. 23626–23636, 2017. DOI: https://doi.org/10.1109/access.2017.2762703.
V. Subbaraju, M. B. Suresh, S. Sundaram, S. Narasimhan. Identifying differences in brain activities and an accurate detection of autism spectrum disorder using resting state functional-magnetic resonance imaging: A spatial filtering approach. Medical Image Analysis, vol. 35, pp. 375–389, 2017. DOI: https://doi.org/10.1016/j.media.2016.08.003.
V. Subbaraju, S. Sundaram, S. Narasimhan. Identification of lateralized compensatory neural activities within the social brain due to autism spectrum disorder in adolescent males. European Journal of Neuroscience, vol. 47, no. 6, pp. 631–642, 2018. DOI: https://doi.org/10.1111/ejn.13634.
H. Dhayne, R. Haque, R. Kilany, Y. Taher. In search of big medical data integration solutions - A comprehensive survey. IEEE Access, vol. 7, pp. 91265–912900, 2019. DOI: https://doi.org/10.1109/ACCESS.2019.2927491.
G. Ariyarathne, S. De Silva, S. Dayarathna, D. Meedeniya, S. Jayarathne. ADHD identification using convolutional neural network with seed-based approach for fMRI data. In Proceedings of the 9th International Conference on Software and Computer Applications, ACM, Langkawi, Malaysia, pp. 31–35, 2020. DOI: https://doi.org/10.1145/3384544.3384552.
X. L. Peng, P. Lin, T. S. Zhang, J. Wang. Extreme learning machine-based classification of ADHD using brain structural MRI data. PLoS One, vol. 8, no. 11, Article number e79476, 2013. DOI: https://doi.org/10.1371/journal.pone.0079476.
D. P. Kuang, X. J. Guo, X. An, Y. L. Zhao, L. H. He. Discrimination of ADHD based on fMRI data with deep belief network. Intelligent Computing in Bioinformatics, D. S. Huang, K. Han, M. Gromiha, Eds., Cham, Switzerland: Springer, pp. 225–232, 2014. DOI: https://doi.org/10.1007/978-3-319-09330-727.
F. X. Castellanos, D. S. Margulies, C. Kelly, L. Q. Uddin, M. Ghaffari, A. Kirsch, D. Shaw, Z. Shehzad, A. Di Martino, B. Biswal, E. J. S. Sonuga-Barke, J. Rotrosen, L. A. Adler, M. P. Milham. Cingulate-precuneus interactions: A new locus of dysfunction in adult attention-deficit/hyperactivity disorder. Biological Psychiatry, vol. 63, no. 3, pp. 332–337, 2008. DOI: https://doi.org/10.1016/j.biopsych.2007.06.025.
C. Fassbender, H. Zhang, W. M. Buzy, C. R. Cortes, D. Mizuiri, L. Beckett, J. B. Schweitzer. A lack of default network suppression is linked to increased distractibility in ADHD. Brain Research, vol. 1273, pp. 114–128, 2009. DOI: https://doi.org/10.1016/j.brainres.2009.02.070.
A. M. S. Aradhya, A. Joglekar, S. Suresh, M. Pratama. Deep transformation method for discriminant analysis of multi-channel resting state fMRI. In Proceedings of the 33rd AAAI Conference on Artifícial Intelligence, AAAI, Hawaii, USA, pp. 2556–2563, 2019. DOI: https://doi.org/10.1609/aaai.v33i01.33012556.
A. M. S. Aradhya, V. Subbaraju, S. Sundaram and N. Sundararajan. Regularized spatial filtering method (RSFM) for detection of attention deficit hyperactivity disorder (ADHD) from resting-state functional magnetic resonance imaging (rs-fMRI). In Proceedings of the 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, IEEE, Honolulu, USA, USA, pp. 5541–5544, 2018. DOI: https://doi.org/10.1109/embc.2018.8513522.
K. Konrad, S. B. Eickhoff. Is the ADHD brain wired differently? A review on structural and functional connectivity in attention deficit hyperactivity disorder Human Brain Mapping, vol. 31, no. 6, pp. 904–916, 2010. DOI: https://doi.org/10.1002/hbm.21058.
I. A. Strigo, S. C. Matthews, A. N. Simmons. Decreased frontal regulation during pain anticipation in unmedicated subjects with major depressive disorder. Translational Psychiatry, vol. 3, no. 3, Article number e239, 2013. DOI: https://doi.org/10.1038/tp.2013.15.
A. J. Hao, B. L. Ha, C. H. Yin. Discrimination of ADHD children based on deep Bayesian network. In Proceedings of IET International Conference on Biomedical Image and Signal Processing, IEEE, Beijing, China, 2015. DOI: https://doi.org/10.1049/cp.2015.0764.
A. Tenev, S. Markovska-Simoska, L. Kocarev, J. Pop-Jordanov, A. Muller, G. Candrian. Machine learning approach for classification of ADHD adults. International Journal of Psychophysiology, vol. 93, no. 1, pp. 162–166, 2014. DOI: https://doi.org/10.1016/j.ijpsycho.2013.01.008.
S. Sartipi, H. Kalbkhani, P. Ghasemzadeh, M. G. Shayesteh. Stockwell transform of time-series of fMRI data for diagnoses of attention deficit hyperactive disorder. Applied Soft Computing, vol. 86, Article number 105905, 2020. DOI: https://doi.org/10.1016/j.asoc.2019.105905.
D. P. Kuang, L. H. He. Classification on ADHD with deep learning. In Proceedings of International Conference on Cloud Computing and Big Data, IEEE, Wuhan, China, pp. 27–32, 2014. DOI: https://doi.org/10.1109/ccbd.2014.42.
G. Deshpande, P. Wang, D. Rangaprakash, B. Wilamowski. Fully connected cascade artificial neural network architecture for attention deficit hyperactivity disorder classification from functional magnetic resonance imaging data. IEEE Transactions on Cybernetics, vol. 45, no. 12, pp. 2668–2679, 2015. DOI: https://doi.org/10.1109/tcyb.2014.2379621.
G. Brihadiswaran, D. Haputhanthri, S. Gunathilaka, D. Meedeniya, S. Jayarathna. EEG-based processing and classification methodologies for autism spectrum disorder: A review. Journal of Computer Science, vol. 15, no. 8, pp. 1161–1183, 2019. DOI: https://doi.org/10.3844/jcssp.2019.1161.1183.
V. Sachnev, S. Suresh, N. Sundararajan, B. S. Mahanand, M. W. Azeem, S. Saraswathi. Multi-region risk-sensitive cognitive ensembler for accurate detection of attention-Deficit/Hyperactivity disorder. Cognitive Computation, vol. 11, no. 4, pp. 545–559, 2019. DOI: https://doi.org/10.1007/s12559-019-09636-0.
M. Delavarian, F. Towhidkhah, P. Dibajnia, S. Gharibzadeh. Designing a decision support system for distinguishing ADHD from similar children behavioral disorders. Journal of Medical Systems, vol. 36, no. 3, pp. 1335–1343, 2010. DOI: https://doi.org/10.1007/s10916-010-9594-9.
K. C. Chu, Y. S. Huang, C. F. Tseng, H. J. Huang, C. H. Wang, H. Y. Tai. Reliability and validity of DS-ADHD: A decision support system on attention deficit hyperactivity disorders. Computer Methods and Programs in Biomedicine, vol. 140, pp. 241–248, 2017. DOI: https://doi.org/10.1016/j.cmpb.2016.12.003.
ADHD-Care, 2019, [Online], Available: http://bloomingsands-73478.herokuapp.com, May 2, 2020.
T. J. Andrews, S. D. Halpern, D. Purves. Correlated size variations in human visual cortex, lateral geniculate nucleus, and optic tract. The Journal of Neuroscience, vol. 17, no. 8, pp. 2859–2868, 1997. DOI: https://doi.org/10.1523/jneurosci.17-08-02859.1997.
X. Nie, Y. Shao, S. Y. Liu, H. J. Li, A. L. Wan, S. Nie, D. C. Peng, X. J. Dai. Functional connectivity of paired default mode network subregions in primary insomnia. Neuropsychiatric Disease and Treatment, vol. 11, pp. 3085–3093, 2015. DOI: https://doi.org/10.2147/ndt.s95224.
V. K. Ha, J. C. Ren, X. Y. Xu, S. Zhao, G. Xie, V. Masero, A. Hussain. Deep learning based single image super-resolution: A survey. International Journal of Automation and Computing, vol. 16, no. 4, pp. 413–426, 2019. DOI: https://doi.org/10.1007/s11633-019-1183-x.
T. Honderich. The Oxford Companion to Philosophy, 2nd ed., Oxford, UK: Oxford University Press, 2005.
I. Zaidi, M. Chtourou, M. Djemel. Robust neural control of discrete time uncertain nonlinear systems using sliding mode backpropagation training algorithm. International Journal of Automation and Computing, vol. 16, no. 2, pp. 213–225, 2017. DOI: https://doi.org/10.1007/s11633-017-1062-2.
S. Ruder. An overview of gradient descent optimization algorithms, [Online], Available: https://arxiv.org/abs/1609.04747, May 2, 2020.
X. H. Zhou, N. A. Obuchowski, D. K. McClish. Statistical Methods in Diagnostic Medicine, 2nd ed., Hoboken, USA: Wiley, 2011.
M. H. Zweig, G. Campbell. Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clinical Chemistry, vol. 39, no. 4, pp. 561–577, 1993. DOI: https://doi.org/10.1093/clinchem/39.4.561.
I. Unal. Defining an optimal cut-point value in ROC analysis: An alternative approach. Computational and Mathematical Methods in Medicine, vol. 2017, Article number 3762651, 2017. DOI: https://doi.org/10.1155/2017/3762651.
K. H. Zou, C. R. Yu, K. Z. Liu, M. O. Carlsson, J. Cabrera. Optimal thresholds by maximizing or minimizing various metrics via ROC-type analysis. Academic Radiology, vol. 20, no. 7, pp. 807–815, 2013. DOI: https://doi.org/10.1016/j.acra.2013.02.004.
R. Fluss, D. Faraggi, B. Reiser. Estimation of the Youden index and its associated cutoff point. Biometrical Journal, vol. 47, no. 4, pp. 458–472, 2005. DOI: https://doi.org/10.1002/bimj.200410135.
N. J. Perkins, E. F. Schisterman. The inconsistency of “optimal” cutpoints obtained using two criteria based on the receiver operating characteristic curve. American Journal of Epidemiology, vol. 163, no. 7, pp. 670–675, 2006. DOI: https://doi.org/10.1093/aje/kwj063.
I. Subramanian, S. Verma, S. Kumar, A. Jere, K. Anamika. Multi-omics data integration, interpretation, and its application. Bioinformatics and Biology Insights, vol. 14, pp. 1–24, 2020. DOI: https://doi.org/10.1177/1177932219899051.
S. V. Faraone, P. Asherson, T. Banaschewski, J. Biederman, J. K. Buitelaar, J. A. Ramos-Quiroga, L. A. Rohde, E. J. S. Sonuga-Barke, R. Tannock, B. Franke. Attention-deficit/hyperactivity disorder. Nature Reviews Disease Primers, vol. 1, Article number 15020, 2015. DOI: https://doi.org/10.1038/nrdp.2015.20.
E. Hoekzema, S. Carmona, J. A. Ramos-Quiroga, V. Richarte Fernandez, R. Bosch, J. C. Soliva, M. Rovira, A. Bulbena, A. Tobena, M. Casas, O. Vilarroya. An independent components and functional connectivity analysis of resting state fMRI data points to neural network dysregulation in adult ADHD. Human Brain Mapping, vol. 35, no. 4, pp. 1261–1272, 2014. DOI: https://doi.org/10.1002/hbm.22250.
Z. Y. Mao, Y. Su, G. Q. Xu, X. P. Wang, Y. Huang, W. H. Yue, L. Sun, N. X. Xiong. Spatio-temporal deep learning method for ADHD fMRI classification. Information Sciences, vol. 499, pp. 1–11, 2019. DOI: https://doi.org/10.1016/j.ins.2019.05.043.
Q. Xu, M. Zhang, Z. H. Gu, G. Pan. Overfitting remedy by sparsifying regularization on fully-connected layers of CNNs. Neurocomputing, vol. 328, pp. 69–74, 2019. DOI: https://doi.org/10.1016/j.neucom.2018.03.080.
J. Sauro. Measuring usability with the system usability scale (SUS): A practical guide to measuring usability, 2019, [Online], Available: https://measuringu.com/sus/, 4, 2019.
A. Riaz, M. Asad, S. M. M. R. Al Arif, E. Alonso, D. Dima, P. Corr, G. Slabaugh. Deep fMRI: AN end-to-end deep network for classification of fMRI data. In Proceedings of the 15th International Symposium on Biomedical Imaging, IEEE, Washington, USA, pp. 1419–1422, 2018. DOI: https://doi.org/10.1109/isbi.2018.8363838.
G. Jayawardena, A. Michalek, S. Jayarathna. Eye tracking area of interest in the context of working memory capacity tasks. In Proceedings of the 20th International Conference on Information Reuse and Integration for Data Science, IEEE, Los Angeles, USA, pp. 208–215, 2019. DOI: https://doi.org/10.1109/iri.2019.00042.
I. Rubasinghe, D. Meedeniya. Ultrasound nerve segmentation using deep probabilistic programming. Journal of ICT Research and Applications, vol. 13, no. 3, pp. 241–256, 2019. DOI: https://doi.org/10.5614/itbj.ict.res.appl.2019.13.3.5.
Z. J. Yao, J. Bi, Y. X. Chen. Applying deep learning to individual and community health monitoring data: A survey. International Journal of Automation and Computing, vol. 15, no. 6, pp. 643–655, 2018. DOI: https://doi.org/10.1007/s11633-018-1136-9.
D. Haputhanthri, G. Brihadiswaran, S. Gunathilaka, D. Meedeniya, S. Jayarathna, M. Jaime, C. Harshaw. Integration of facial thermography in EEG-based classification of ASD. International Journal of Automation and Computing, to be published, vol. 17, no. 6, pp. 837–854 DOI: https://doi.org/10.1007/s11633-020-1231-6.
Acknowledgements
This work was supported by Old Dominion University, Norfolk, Virginia, USA and University of Moratuwa, Sri Lanka. We thank the participants of the system usability study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Colored figures are available in the online version at https://link.springer.com/journal/11633
Senuri De Silva is a bachelor student in computer science and engineering at Department of Computer Science and Engineering, University of Moratuwa, Sri Lanka.
Her research interests include biomedical, machine learning and data mining.
Sanuwani Dayarathna is a bachelor student in computer science and engineering at Department of Computer Science and Engineering, University of Moratuwa, Sri Lanka.
Her research interests include biomedical, machine learning and data mining.
Gangani Ariyarathne is a bachelor student in computer science and engineering at Department of Computer Science and Engineering, University of Moratuwa, Sri Lanka.
Her research interests include machine learning, data mining, biomedical, and computer security.
Dulani Meedeniya received the Ph.D. degree in computer science from University of St Andrews, UK in 2013. She is a senior lecturer in Department of Computer Science and Engineering, University of Moratuwa, Sri Lanka. She is a Fellow of Higher Education Academy, UK, Member of The Institution of Engineering and Technology, Member of Institute of Electrical and Electronics Engineers, and a Charted Engineer registered at Engineering Council, UK.
Her research interests include software modelling and design, workflow tool support for bioinformatics, data visualization and recommender systems.
Sampath Jayarathna received the Ph. D. degree in computer science from the Texas A&M University - College Station, USA in 2016. He is an assistant professor in computer science at Old Dominion University, USA, where he is associated with Web Science and Digital Libraries (WS-DL) research group. He is a member of Association for Computing Machinery (ACM), IEEE, and Scientific Research Honor Society (Sigma XI).
His research interests include machine learning, information retrieval, data science, eye tracking, and brain-computer interfacing.
Anne M. P. Michalek received the Ph.D. degree in special education from Old Dominion University, USA in 2012. She is an associate professor of communication disorders and special education at Old Dominion University, USA.
Her research interests include multi-disciplinary approaches using biomedical technologies and instructional tools to improve outcomes for at-risk students and students with ADHD and autism spectrum disorder (ASD).
Rights and permissions
About this article
Cite this article
De Silva, S., Dayarathna, S., Ariyarathne, G. et al. Computational Decision Support System for ADHD Identification. Int. J. Autom. Comput. 18, 233–255 (2021). https://doi.org/10.1007/s11633-020-1252-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11633-020-1252-1