Abstract
Fungal specimens parasitic on spider egg sacs (Araneidae sensu lato) were collected, isolated, and identified based on molecular phylogenetic analyses of five nuclear loci (ITS, LSU, TEF1, RPB1 and RPB2) combined with morphological data. In this study, one novel monotypic genus is described, Bhushaniella rubra for Thailand. Bhushaniella rubra is characterized by producing superficial perithecia. Its anamorph has a unique character by producing verticillate phialides with a slightly curved neck. A concurrent evaluation of the secondary metabolites of the mycelial extracts of the new fungus revealed the presence of picoline alkaloids of the penicolinate type, for which we propose the trivial names penicolinates F and G. Their chemical structures were elucidated by two-dimensional nuclear magnetic resonance (2D-NMR) spectroscopy and high resolution mass spectrometry (HR-MS). They only showed weak to no antibiotic activity and were devoid of significant cytotoxic effects.
Similar content being viewed by others
Introduction
Cordycipitaceae Kreisel ex G.H. Sung, J.M. Sung, Hywel-Jones & Spatafora (Hypocreales, Ascomycota) is a heterogeneous family containing species with diverse morphological characteristics of their stromata. The diversity of these characters range from having a soft, fleshy texture and pallid (white to yellow) to brightly coloured (orange to red) stipitate stromata with loosely embedded or superficial perithecia e.g. Blackwellomyces cardinalis (G.H. Sung & Spatafora) Spatafora & Luangsa-ard on Lepidoptera larvae, Beauveria mimosiformis Khons., Thanakitp., Kobmoo & Luangsa-ard on Coleoptera, Samsoniella inthanonensis Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard on Lepidoptera larvae (Sung and Spatafora 2004; Mongkolsamrit et al. 2018; Khonsanit et al. 2020), to possessing non-stipitate ascomata such as species in Hyperdermium J.F. White, R.F. Sullivan, Bills & Hywel-Jones and Neohyperdermium Thanakitp. & Luangsa-ard (Thanakitpipattana et al. 2022).
Previous studies employing molecular phylogenetics revealed that members of Cordycipitaceae occur on a wide range of hosts and substrates. They are well-known attacking multiple orders of insects from larvae to adult states and spiders. Some species in Beauveria Vuill. and Cordyceps Fr. could be isolated from soil or be established as endophytes in plant seedlings (Zimmermann 2008; Alali et al. 2019; Ramakuwela et al. 2020). Furthermore, there is evidence that some species in this family like Simplicillium lanosoniveum (J.F.H. Beyma) Zare & W. Gams and Niveomyces coronatus J.P.M. Araújo & de Bekker are mycoparasites of Ophiocordyceps species pathogenic on ants. (Shrestha et al. 2016, 2019; Wei et al 2019; Araújo et al. 2022). Many studies have shown that some species in Cordycipitaceae have important economic value being used as biocontrol agents for agricultural insect pests or are sources of bioactive compounds. For instance, Beauveria bassiana (Bals.-Criv.) Vuill. can be used as a biocontrol agent to reduce the population of Odoiporus longicollis Olivier, which has a severe impact on banana production (Alagesan et al. 2019). Strains of Cordyceps fumosorosea (Wize) Kepler, B. Shrestha & Spatafora (= Isaria fumosorosea Wize) are frequently used for whitefly control (Avery et al. 2004; 2008). In addition, several species in Cordycipitaceae produce secondary metabolites with bioactivities that have the potential for medicines or nutriment e.g. C. militaris (L.) Fr., C. cicadae (Miq.) Massee, and C. tenuipes (Peck) Kepler, B. Shrestha & Spatafora (Zhang et al. 2018; Jędrejko et al. 2021). Two bioactive compounds, gibellamines A (1) and B (2) were recently isolated from Gibellula gamsii Kuephadungphan, Tasan. & Luangsa-ard showing anti-biofilm activity against Staphylococcus aureus (Kuephadungphan et al. 2019). The exploration of bioactive compounds from species of invertebrate-pathogenic fungi has recently received increasing interest (Helaly et al. 2019; Zhang et al. 2020; Mongkolsamrit et al. 2021).
In surveys of arthropod pathogenic fungi in Thailand’s national parks, collections of pathogens on spider egg sacs were found on the underside of leaves of forest plants. Based on the macroscopic features of the teleomorph, specimens possess superficial perithecia on the spider eggs in a sac, which is similar to the teleomorph in Gibellula. Nonetheless, asexually reproductive species produce cylindrical synnema with verticillate phialides along the synnema and on hosts. These studies aim to elucidate the phylogenetic and taxonomic placement of these collections of parasitic fungi on spider eggs in a sac through multilocus molecular phylogenetic analyses to known members of Cordycipitaceae and an investigation of the bioactivity of secondary metabolites produced by these fungi is presented.
Materials and methods
Specimen collection and isolation
The fungal specimens were collected from different forests in Thailand, located in Chumphon Province. They were searched on the underside of leaves and placed in plastic boxes. The protocol for isolating ascospores and conidia followed a previous study (Mongkolsamrit et al. 2018) using potato dextrose agar (PDA) plates (PDA: freshly diced potato 200 g/L, dextrose 20 g/L, agar 15 g/L). After the inoculated medium was incubated overnight at room temperature, it was examined with a stereomicroscope to locate germinated ascospores and conidia. The germinated ascospores and conidia were transferred to fresh PDA plated and then incubated for 14 days at 25 °C under light/dark conditions (L:D 14:10). The cultures were deposited at the BIOTEC Culture Collection (BCC), National Center for Genetic Engineering and Biotechnology, Thailand. All fungal specimens were dried in an electric food dryer (50–55 °C) overnight and accessioned in the BIOTEC Bangkok Herbarium (BBH), National Biobank of Thailand.
Morphological observation
Macroscopic characters were observed based on natural specimens and pure cultures. Microscopic characters of perithecia, asci, ascospores, phialides and conidia were mounted on a microscope slide containing a drop of lactophenol cotton blue solution. The shapes, sizes, and colours of individual characters were determined and measured according to Mongkolsamrit et al. (2020). Fungal strains were grown on oatmeal agar (OA, Difco: oatmeal 60 g/L, agar 12.5 g/L) and PDA agar plates at 25 °C under light/dark conditions (L:D 14:10) for 14 days. The cultures were observed to compare morphological characters including conidia, phialides, and colony pigmentation. The colours of fresh specimens and cultures incubated on OA and PDA were described and codified following the Royal Horticultural Society colour chart (RHS Colour Chart 2015).
Molecular phylogenetic analyses
Genomic DNA was harvested from mycelial mass on PDA using a modified cetyltrimethyl-ammonium bromide (CTAB)(Doyle and Doyle 1987) as previously described by Mongkolsamrit et al. (2020). Nuclear loci, including the nuc rDNA region encompassing the internal transcribed spacer (ITS) regions ITS1 and ITS2, nc LSU rDNA (large subunit of the ribosomal DNA), the translation elongation factor-1α (TEF1), and the partial gene regions of the largest and second-largest subunits of the RNA polymerase II (RPB1 and RPB2), were amplified and sequenced. The primer pairs and thermocycler conditions for PCR amplifications used in this study followed Mongkolsamrit et al. (2023) and Thanakitpipattana et al. (2022). The purified PCR products were sequenced with the same PCR amplification primers for Sanger dideoxy sequencing. The DNA sequences generated in this study were checked for ambiguous bases using BioEdit v. 7.2.5 (Hall 1999) and then submitted to GenBank. Table 1 shows the list of ITS, LSU, TEF1, RPB1 and RPB2 sequences generated in this study as well as those of other taxa from previous studies. Phylogenetic analyses were performed using RAxMLHPC2 on XSEDE v 8.2.12 (Stamatakis 2014) in the CIPRES Science Gateway portal, using the GTRGAMMA + I model with 1000 bootstrap iterations. Bayesian inference (BI) of phylogenetic relationships was performed in MrBayes v. 3.2.7a (Ronquist et al. 2012), with best-fit models selected using MrModeltest v. 2.2 (Nylander 2004). The best model was GTR + G + I. Markov chain Monte Carlo (MCMC) simulations were run for 5,000,000 generations, sampling every 1000 and discarding the first 10% as burn-in. RAxML and BI output were imported into TreeView version 1.6.6 to visualize the phylogenetic trees (Page 1996).
Instrumentation for spectral measurements
NMR spectra were recorded on a Bruker Avance III 700 spectrometer with a 5 mm TXI cryoprobe (1H 700 MHz, 13C 175 MHz) and a Bruker Avance III 500 (1H 500 MHz, 13C 125 MHz) spectrometer and referenced to the adopted solvent peaks. HPLC–MS analyses were performed on a Dionex UltiMate 3000 UHPLC (Thermo Fisher Scientific Inc., Waltham, MA, USA) with a diode array detector and C18 Acquity UPLC BEH column (2.1 × 50 mm, 1.7 μm, Waters, Eschborn, Germany) with a gradient previously published (Schrey et al. 2022); connected to an ion trap ESI–MS (amaZon speed™, Bruker Daltonics, Bremen, Germany). HR-ESI–MS spectra were measured on a time-of-flight (TOF) MS (maXis II™, Bruker Daltonics). All chemicals and solvents were acquired from AppliChem GmbH (Darmstadt, Germany), Avantor Performance Materials (Deventor, The Netherlands), Carl Roth GmbH & Co. KG (Karlsruhe, Germany), or Merck KGaA (Darmstadt, Germany) in analytical and HPLC grade.
Fermentation and Extraction
For 4 L fermentation, twenty 500 mL Erlenmeyer flasks, each containing 200 mL YMG media (4 g yeast extract; 10 g malt extract; 4 g D-glucose; ad 1000 ml distilled water) were inoculated with 7 mycelial plugs (approx. 1 cm × 1 cm) from an actively growing colony of strain BCC 47541. After incubation at 23 °C on a rotary shaker (140 rpm) for 7 to 8 days, until free glucose was depleted in every flask, the culture broth was separated from the mycelia by vacuum filtration and the mycelia subsequently extracted with acetone, followed by EtOAc to afford roughly 300 mg of mycelial extract as brown gum.
Isolation of Metabolites 1 and 2
The extract was separated in 3 runs through a Kromasil RP C18 column (250 mm × 20 mm, 7 µm, MZ-Analysentechnik, Mainz, Germany) using deionized water (Milli-Q Millipore) and acetonitrile (HPLC grade) as the mobile phase on an Agilent 1100 series HPLC system (Agilent Technologies, Wilmington, DE, USA). The separation was carried out according to the following gradient: from 20% – 75% acetonitrile in 50 min, then rising to 100% ACN in 10 min, and maintaining 100% for 5 min. UV detection was performed at 220, 280, and 325 nm. Fractions were collected and pooled according to the observed peaks. The separation yielded two fractions including compound 1 (1.2 mg) and 2 (1.1 mg) at retention times tR 15–16 and 28–29 min, respectively.
Biological assays
The minimum inhibitory concentrations (MICs) of compounds 1 and 2 were determined in a serial dilution assay for Rhodotorula glutinis DSM10134, Staphylococcus aureus DSM346, Candida albicans DSM1665, Mycobacterium smegmatis ATCC700084 and Mucor hiemalis DSM2656 and cytotoxicity was tested against the murine fibroblast (L929) human HeLa (KB3.1), human adenocarcinoma (MCF-7), human epidermoid carcinoma (A431), adenocarcinomic human alveolar basal epithelial (A549), and human prostate carcinoma (PC-3) cell lines, as described by Sandargo et al. (2021).
Results
Molecular phylogeny
We generated 14 new sequences (3 ITS, 3 LSU, 3 TEF1, 3 RPB1 and 2 RPB2) from living cultures (Table 1). Purpureocillium lilacinum (CBS 431.87 and CBS 284.36) was used as outgroup. The combined dataset from 53 specimens, with multi-locus sequences totaling an alignment length of 4,040 characters with gaps (ITS 653, LSU 840, TEF1 909, RPB1 750 and RPB2 888) was analysed. The maximum-likelihood phylogenetic analyses resulted in a multi-locus tree with maximum likelihood bootstrap values (MLB) shown in Fig. 1. The nodes were also evaluated with Bayesian posterior probabilities (BPP).
The phylogenetic analyses supported three strains BCC 47515, BCC 47541 and BCC 47542 that group together as a monophyletic clade with maximum support (MLB = 100/ BPP = 1), branched as sister to the four genera occurring on spiders and spider eggs in a sac including Gibellula, Hevansia, Jenniferia and Polystromomyces, and is thus proposed as a new genus Bhushaniella which contains a new species, Bhushaniella rubra. The sequence alignments for all datasets used in this study are provided in 106084/m9.figshare.22810688.v1.
Taxonomy
Bhushaniella Mongkolsamrit Noisripoom & Luangsa-ard, gen. nov.
MycoBank No: 849077.
Type species: Bhushaniella rubra Mongkolsamrit, Noisripoom & Luangsa-ard.
Etymology: In honour of Dr. Bhushan Shrestha for his contribution to the knowledge of arthropod-pathogenic fungi.
Description: Spider eggs in a sac are covered with pale yellowish white to moderate yellow mycelium. Teleomorph: Perithecia produced on the mycelial mat covering the body of the hosts, superficial with mycelia covering the bottom half of the perithecium, ovoid narrowing towards the ostiole. Asci cylindrical with thickened caps, 8-spored. Ascospores filiform, hyaline, whole. Anamorph: Synnema produced from the mycelial mat-covering hosts, erect, unbranched, solitary, and cylindrical. Conidiophores erect arising along with the synnema, occasionally found on the mycelium covering the hosts, verticillate with phialides in whorls of two to five. Phialides comprising a cylindrical basal portion, tapering into a thin slightly curved neck. Conidia fusiform, slightly curved, aggregated at the apex of the phialides.
Notes: Bhushaniella contains one species, Bh. rubra. The teleomorph state is morphologically similar to Gibellula spp. by producing superficial perithecia on the mycelial mat covering the body of the hosts. However, it differs from Gibellula spp. in producing whole ascospores. The ascospores of Gibellula spp. are multiseptate and disarticulate. Additionally, the anamorph in Bhushaniella produces verticillate phialides with a slightly curved neck.
Bhushaniella rubra Mongkolsamrit, Noisripoom & Luangsa-ard, sp. nov. (Fig. 2).
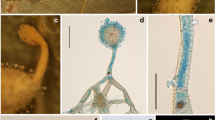
Bhushaniella rubra. a Fungus arising from spider eggs in a sac (BBH 31245). b Perithecium c Asci d Ascus tip e Filiform whole ascospores in ascus. f Fungus on a spider egg sac (BBH 31241). g Conidiogenous cells forming verticillated phialides. h–i Phialides with conidia. j–k Culture characters on OA with reddish pigment diffusing in agar medium (j obverse, k reverse). l-m Phialide apex with conidial head on OA. n Conidia on OA. o–p Microcycle conidiation on OA. q–r Culture characters on PDA with reddish pigment diffusing in agar medium (q obverse, r reverse). s–t Apices of phialides apex with conidial heads on PDA. u Conidia on PDA. v–w Microcycle conidiation on PDA Scale bars: a, f = 2 mm, b = 300 µm, c = 100 µm, g, m, s, t = 20 µm, d, e = 5 µm, h, i, l, n, o, p, u, v, w = 10 µm
MycoBank No: 849078.
Holotype: Thailand, Chumphon Province, Phato Watershed Conservation and Management Unit, on spider eggs sac (Araneidae sensu lato) attached to the underside of a palm leaf, 10 March 2010, K. Tasanathai, P. Srikitikulchai, A. Khonsanit, K. Sansatchanon, D. Thanakitpipattana, MY6567.01 (BBH 31245, holotype), ex-type culture BCC 47541 isolated from ascospores.
Etymology: The name refers to the pale red pigment produced by the fungus when growing on PDA and OA.
Description: Spider eggs sac are covered with pale yellowish white (155 D) to moderate yellow mycelium (161 A-B). Teleomorph: Perithecia produced on the mycelial mat covering the body of the hosts, superficial with mycelia covering the bottom half of the perithecium, ovoid narrowing towards the ostiole, (500–)538–640(–660) × (280–)300–360(–380) µm. Asci cylindrical, (100–)250–380(–400) × 3–5 µm, with cap 2–3 μm thick. Ascospores hyaline, filiform, whole ascospores, extending the length of ascus. Anamorph: Synnema produced from the host, erect, unbranched, solitary, cylindrical, 5–8 mm long, ca. 0.5 mm wide. Conidiophores produced along the synnema, on the mycelium covering the hosts. Phialides consisting of a cylindrical basal portion, verticillate, with phialides in whorls of two to five, occasionally with solitary phialides, (8–)9–14(–15) × 1–2.5 µm µm, tapering into a distinct slightly curved neck, (3–)4.5–7.5(–8) × 1–1.5 µm. Conidia fusiform, slightly curved, aggregated at the apex of the phialides, 4–5 × 1–2 µm.
Culture characteristics: Colonies on OA attaining a diam. of 8–10 mm in 14 days, cottony with high mycelium density, white, reverse moderate yellowish pink (N170C), producing reddish pigment diffusing in agar medium. Phialides arising from vegetative hyphae, solitary, narrow cylindrical basal portion, 10–25.5(–35) × 1–2 µm. Conidia hyaline, smooth, aggregated at the apex of the phialides, distinctly of two types: 1) cylindrical, aseptate, (3–)4.5–7.5(–8) × (1–)1.5–2.5 µm; 2) fusiform, early in development aseptate, producing 1 septum, (8–)8.5–14.5(–18) × 2–3 µm. Conidial arrangement acremonium-like. Microcycle conidiation observed, cylindrical, 2–4(–5) × 1–2 µm.
Colonies on PDA attaining a diam. of 10–15 mm in 14 days, cottony with high mycelium density, white, reverse moderate yellowish pink (N170C), producing reddish pigment diffusing in agar medium. Phialides arising from vegetative hyphae, solitary, narrow cylindrical basal portion, (10–)12.5–35(–50) × 0.5–2 µm. Conidia hyaline, smooth, aggregated at the apex of the phialides, distinctly of two types: 1) cylindrical, aseptate, (3–)5–7.5(–8) × 1.5–2 µm; 2) fusiform, early in development aseptate, becoming 1 septum, 10–15.5(–18) × 2–3 µm, aggregated at the apex of the phialides. Conidial arrangement acremonium-like. Microcycle conidiation observed, cylindrical (2–)2.5–4(–5) × 1–2 µm.
Distribution: Found in southern Thailand.
Additional materials examined: Thailand, Chumphon Province, Phato Watershed Conservation and Management Unit, spider eggs sac (Araneidae sensu lato) attached to the underside of a dicot leaf of forest plants, 10 March 2010, K. Tasanathai, P. Srikitikulchai, A. Khonsanit, K. Sansatchanon, D. Thanakitpipattana, MY6567.02 (BBH 31245, paratype), ex-paratype culture BCC 47542 isolated from conidia, idem, MY6566.01 (BBH 31244), culture BCC 47539 isolated from ascospores, idem, MY6566.02 (BBH 31244), culture BCC 47540 isolated from conidia. Chumphon Province, Heo Lom Waterfall, spider eggs in a sac (Araneidae sensu lato) attached to the underside of a dicot leaf of forest plants, 9 March 2010, K. Tasanathai, P. Srikitikulchai, A. Khonsanit, K. Sansatchanon, D. Thanakitpipattana, MY06532 (BBH 31241), culture BCC 47515 isolated from conidia.
Notes. Bhushaniella rubra shows microcycle conidiation from conidia on culture (OA and PDA), it has also been observed in Po. araneae, which occurs on spiders egg sacs as well. Bhushaniella rubra produces a reddish pigment diffusing in OA and PDA plates, while Po. araneae does not produce any pigment in agar medium.
Secondary metabolites
Compound 1 was isolated as a colourless solid that revealed pseudomolecular ion peak at m/z 385.2 [M + H]+ suggesting the molecular weight to be of 384 g/mol. Its molecular formula was established to be C22H28N2O4 based on HRESIMS that exhibited a pseudomolecular ion peak at m/z 385.2120 [M + H]+ (calculated for C22H29N2O4; 385.2127) indicating ten degrees of unsaturation. The UV spectrum of 1 displayed three maximal absorption peaks (λmax) at 199, 226 and 273 nm. The 13C NMR spectrum of 1 (Table 2, see supplementary materials Figure S7) showed the presence of ten carbon resonances suggesting that compound 1 is either a dimer of two identical monomers or a symmetric compound with each carbon peak represents two electromagnetically equivalent carbon atoms in 1. The ten carbon resonances can be recognized into one carbonyl carbon (δc 166.1) and two quaternary carbons (δC 146.0, 141.7), three olefinic carbons (δC 149.4, 137.0, 124.4) along with five methylene carbons (δC 32.0, 30.3, 28.9, 28.7, 28.5). The 1H NMR and HSQC spectra of 1 (Table 2, see supplementary materials Figures S6 and S10) revealed three deshielded olefinic protons at δH 8.54 (d, J = 2.2 Hz), δH 7.96 (dd, J = 8.0 Hz) and δH 7.80 (d, J = 8.0, 2.2 Hz) that are directly correlated to three olefinic carbons at δc 149.4, 124.4 and 137.0, respectively. These results indicated the presence of a 2,5-disubstituted pyridine moiety that by searching the available literature suggested that compound 1 is structurally related to picolinic acid fungal metabolites, penicolinates A-C (Intaraudom et al. 2013). By comparing the MS and NMR spectral data between metabolite 1 and penicolinate A (Intaraudom et al. 2013), it was obviously that 1 has less molecular weight by 28 amu compared to penicolinate A that has been reflected in 1H NMR by the absence of methyl ester group at δH 3.97 ppm and the appearance of a highly deshielded broad proton resonance at δH 13.00 ppm assigned to the free carboxylic acid group. The aforementioned results suggested that compound 1 to be the free carboxylic acid derivative of penicolinate A featuring a symmetrical compound comprising two 2,5-disubstituted pyridine moieties bonded through a ten-methylene chain. Further confirmation of the ascribed chemical structure of 1 was provided by 2D NMR spectra including 1H-1H-COSY and HMBC (Table 2, Fig. 4, see supplementary materials Figure S5, S6). The 1H-1H-COSY (Table 2, Figs. 3 and 4) revealed two main spin systems, one between three olefinic protons at δH 8.54, δH 7.96 and δH 7.80 assigned to H-6, H-3 and H-4, respectively, whereas the second spin system was among the five methylene protons at δH 2.65 (t, J = 7.7 Hz), δH 1.60 (quin, J = 7.7 Hz) and three methylenes at δH 1.15–1.35 (m). In addition, a long-range COSY correlations (Table 2, Fig. 4) were also noticed between olefinic protons at δH 8.54 and δH 7.80 with the methylene protons at δH 2.65 (t, J = 7.7 Hz) ascribed for H-4, H-6 and CH2-8, respectively. The HMBC spectrum (Table 2, Fig. 4) further confirmed the positions of the carboxylic acid moiety at C-2 via key correlations from H-3 (δH 7.96, dd, J = 8.0 Hz) to the carboxylic acid carbon (δC 166.1) along with the key correlations from H-4 and H-6 to the methylene carbon at (δC 32.0) ascribed to C-8. In conclusion, compound 1 was unambiguously determined to be a new symmetric picolinic acid derivative that was given a trivial name, penicolinate F.
Compound 2 was obtained as an off-white amorphous solid that revealed maximal absorption peaks (λmax) at 200, 218 and 273 nm as those recognized for 1. Its HRESIMS exhibited a pseudomolecular ion peak at m/z 320.1 [M + H]+ suggesting its molecular weight to be of an odd value (319.1 g/mol) and hence supporting the presence of an odd number of nitrogen atoms. This notion was further proved by HRESIMS that displayed a pseudomolecular ion peak at m/z 320.1860 [M + H]+ (calculated for; 320.1862) indicating the molecular formula of 2 to be C18H25NO4 suggesting its inclusion of seven degrees of unsaturation. The 1H, 13C NMR and HSQC spectra of 2 (Table 3, see supplementary materials Figure S13, S14, S18) confirmed the presence of a 2,5-disubstituted pyridine moiety similar to that in 1 by the presence of three olefinic protons at δH 8.53 (d, J = 1.9 Hz), δH 7.95 (d, J = 7.9 Hz) and δH 7.78 (dd, J = 7.9, 1.9 Hz) which directly correlated to three olefinic carbons at δC 149.2, 123.4 and 136.6, respectively. In addition, two olefinic protons at δH 6.95 (dt, J = 15.7, 7.0 Hz) and δH 5.75 (dd, J = 15.7, 1.7 Hz) directly correlated to two olefinic carbons at δC 148.6 and 121.7, respectively, indicating the presence of a trans-α,β-unsaturated carbonyl carbon moiety. Based on the obtained results and by comparison with the reported literature, metabolite 2 was suggested to possess a single picolinic acid moiety, resembling penicolinates D and E (Intaraudom et al. 2013). To further determine the structural characteristics of 2, 2D NMR spectral analyses were conducted including 1H-1H-COSY and HMBC (Table 3, Fig. 4, see supplementary materials Figure S16, S17). The 1H-1H-COSY spectrum of 2 revealed two clear spin systems: one of them deshielded olefinic protons at δH 8.53, δH 7.95 and δH 7.78 ppm proving the presence of a picolinic acid moiety with long-range COSY correlations from the two olefinic protons at δH 8.53 and δH 7.78 to the methylene moieties at δH 2.66 (t, J = 7.7, 2H) and thereafter to δH 1.59 (p, J = 7.5 Hz) and δH 1.17–1.31 (m). The second spin system was starting from the trans-α,β-unsaturated olefinic protons at δH 5.75 and δH 6.95 then extending to the methylene protons at δH 2.15 (m, 2H), δH 1.38 (m, 2H) and further to 1.17–1.31 (m). The HMBC spectrum of 2 (Table 3, Fig. 4) unraveled key long correlations from the two olefinic protons at δH 5.75 and δH 6.95 to a carboxycarbonyl group at δC 167.2 indicating that compound 2 has a single picolinic acid ring linked through nine methylene carbon chain to a trans-α,β-unsaturated carboxylic acid moiety. HMBC spectrum of 2 (Table 3, Fig. 4) further confirmed the binding of the picolinic acid functionality at C-5 to the aliphatic side chain by the key correlations from methylene groups at δH 2.66 (CH2-8) and δH 1.59 (CH2-8) to the carbon resonances at δC 136.6 and δC 143.9 assigned to C-4 and C-5, respectively. Based on the obtained results, compound 2 was confirmed to be an asymmetric picolinic acid derivative related to the previously reported penicolinates D and E, thus it was trivially named as penicolinate G.
Both secondary metabolites did not display any noticable signs of cytotoxicity in the conducted assay. Penicolinate F (1) displayed a minimum inhibitory concentration against Mucor hiemalis at 66.6 µg/ml and penicolinate G (2) against Bacillus subtilis at 66.6 µg/ml. No further inhibitory effects were observed in the conducted assays.
Penicolinate F (1): Colourless solid; UV (MeOH) λmax 199, 226 and 273 nm; 1H and 13C NMR see Table 2; HRESIMS m/z 385.2120 [M + H]+ (calcd for C22H29N2O4; 385.2127).
Penicolinate G (2): Off-white amorphous solid; UV (MeOH) λmax 200, 218 and 273 nm; 1H and 13C NMR see Table 3; HRESIMS m/z 320.1860 [M + H]+ (calcd for C18H26NO4; 320.1862).
Discussion
Based on the evidence of morphological and molecular data from previous studies, it revealed that spider-pathogenic fungi in Cordycipitaceae are present in several genera including Akanthomyces Lebert, Beauveria Vuill., Cordyceps Fr., Engyodontium de Hoog, Gibellula Cavara, Hevansia Luangsa-ard, Hywel-Jones & Spatafora, Jenniferia Mongkols., Noisrip. & Tasan, Lecanicillium W. Gams & Zare, Parahevansia (Hywel-Jones) Mongkols. & Noisrip., Polystromomyces Mongkols., Noisrip., Sakolrak & Himaman, Samsoniella Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard Gamszarea Z.F. Zhang & L. Cai, and Simplicillium W. Gams & Zare. (Kepler et al. 2017; Mongkolsamrit et al. 2018, 2022; Zhang et al. 2021; Chen et al. 2022a, b). In addition to these genera, we propose a new genus with one species (Bhushaniella rubra) which constitutes a strongly supported monophyletic clade (MLB = 100/ BPP = 1 in Fig. 1). From our phylogenetic analyses and morphological comparisons, Bhushaniella is closely related to Hevansia, Jenniferia, Gibellula and Polystromomyces (Fig. 1). Bhushaniella rubra is characterized by producing astipitate ascomata (Fig. 2 a) that share common teleomorph morphological features resembling Gibellula, Jenniferia, spider-pathogenic Akanthomyces e.g. A. thailandicus Mongkols., Spatafora & Luangsa-ard and A. sulphureus Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard which are linked with torrubiella-like teleomorph mainly by the production of superficial perithecia. Jenniferia mainly differs in possessing perithecia the subiculum aggregated in clusters. On the other hand, Hevansia and Polystromomyces share similar morphological characters by producing stipitate ascomata with fertile heads at the terminal end of stipes and perithecia are immersed. The species in these genera are associated with spiders and spider eggs sac, which can be found in the same ecological habitat (on the underside of forest plants) (Mongkolsamrit et al. 2018; 2022; Kuephadungphan et al. 2020; 2022; Mendes-Pereira et al. 2023). Meanwhile, the teleomorph of Cordyceps on spiders normally produces stromata arising from hosts with the fertile parts being the upper part of stromata. For example, C. araneae Mongkols., Tasan., Noisrip., Himaman & Luangsa-ard, C. kuiburiensis Himaman, Mongkols., Noisrip. & Luangsa-ard and C. nidus T. Sanjuan, Chir.-Salom. & S. Restrepo. They can often be found on the ground or leaf litter (Chiriví et al. 2017; Crous et al. 2019; Mongkolsamrit et al. 2020).
Based on the anamorph in the natural specimen, Bh. rubra has a solitary cylindrical synnema. The conidiophores in Bh. rubra are produced along the synnema and on the mycelium covering the hosts. Its phialides consist of a cylindrical basal portion, mostly verticillate and in whorls of two to five. Hence, its morphological characters clearly differ from those of several species in Hevansia, Jenniferia and Gibellula that produce multiple synnemata. Significantly, species in Gibellula produce the synnemata bearing predominantly aspergillus-like conidiophores or occasionally growing penicillate or granulomanus-like conidiophores. The anamorphs of Hevansia and Jenniferia are characterized by producing phialides with mono- or polyphialidic conidiogenous cells with cylindrical or swollen basal portions, and their conidia are not catenulate. Polystromomyces, the anamorph has not been seen in the field. (Kuephadungphan et al. 2020, 2022; Mongkolsamrit et al. 2022).
Beauveria is one of the best-known entomopathogenic fungi with a global distribution (Khonsanit et al. 2020). Samsoniella was established by Mongkolsamrit et al. (2018), there are currently 29 species (Chen et al. 2020; Chen et al. 2021a, b, 2022a; Wang et al. 2020, 2022) that have been recorded in the Index Fungorum (accessed in May 2023). Beauveria and Samsoniella have a wide host range, but were rarely reported from spiders. Currently, Beauveria araneola Wan H. Chen, Y.F. Han, Z.Q. Liang & D.C. Jin and S. farinosa Hong Yu bis, Yao Wang & Z.Q. Wang were found on spiders. Engyodontium rectidentatum (Matsush.) W. Gams, de Hoog, Samson & H.C. Evans is commonly isolated from soil, but can also be found on spiders (Meta menardi Latreille) in the Czech Republic (Gams et al. 1984; Kubátová 2017). Based on the morphological characters, the three aforementioned species produce white mycelium on spiders.
Bhushaniella rubra produces conidiophores bearing whorls of phialides, verticillate or solitary; conidia that are aggregated at the apex of the phialides, corresponding to what is described as the conidiogenesis in Lecanicillium Zare & Gams (2001). Lecanicillium Zare & Gams was segregated from Verticillium sect. Prostrata together with Simplicillium W. Gams & Zare, Pochonia Bat. & O.M. Fonseca, Haptocillium W. Gams & Zare, and Rotiferophthora G.L. Barron (Gams and Zare 2001). Currently, thirty-six species in Lecanicillium have been formally described and recorded in Index Fungorum (May 2023), and all these species are mostly associated with insects, spiders, plants, decayed wood, and soil. The phylogenetic studies confirmed that Lecanicillium is polyphyletic (Kepler et al. 2017; Zhou et al. 2022). Lecanicillium is typified by Lecanicillium lecanii (Zimm.) Zare & W. Gams with Torrubiella confragosa Mains as the corresponding teleomorph and was transferred to Akanthomyces including five species of Lecanicillium (Kepler et al. 2017; Shrestha et al. 2019). Subsequently, Wang et al. (2020) transferred three species of Lecanicillium to Flavocillium H. Yu, Y.B. Wang, Y. Wang, Q. Fan & Zhu L. Yang. Zhang et al. (2020) transferred four species of Lecanicillium with L. wallacei (H.C. Evans) H.C. Evans & Zare (teleomorphic synonym: Torrubiella wallacei H.C. Evans) and Verticillium indonesiacum Kurihara & Sukarno which were found on spiders, to Gamszarea. Furthermore, in the our molecular phylogenetic tree, Bh. rubra formed an independent lineage and did not cluster with two species, e. g., Le. tenuipes (Petch) Zare & W. Gams and Le. aranearum (Petch) Zare & W. Gams occurring on spiders.
Based on our spider-pathogenic fungi collection, we have noted that juveniles and adults of spiders are mainly parasitized by fungi and are rarely found on eggs sac (Kuephadungphan et al. 2020, 2022; Mongkolsamrit et al. 2022). Based on the specimens in this study, the eggs sac was completely covered by mycelium, leading to difficulty in identifying the hosts. Thus far, two species of fungi occurring on eggs sacs of spiders were proposed from Thailand, Bh. rubra (this study) and P. araneae Mongkols., Noisrip., Sakolrak & Himaman (Mongkolsamrit et al. 2022). The multi-locus phylogenetic analyses showed that these two genera are closely related. Additionally, both species produce a microcycle conidiation on agar media (OA and PDA). There are numerous reports of microcycle conidiation from entomopathogenic fungi under laboratory conditions such as B. bassiana and Metarhizium acridum (Driver & Milner) J.F. Bisch., S.A. Rehner & Humber (Bosch and Yantorno 1999; Wang et al. 2016; Song et al 2019). Microcycle conidiation is generally induced under unfavourable conditions. This phenomenon has been discussed to be a mechanism for increasing conidia numbers to increase the ability of the fungi to infect their host and their tolerance to unfavorable environmental conditions (Nishi et al. 2021; Zou et al. 2022).
The ability of Bh. rubra BCC 47541 to produce penicolinates was also found in Bh. rubra BCC 47542 (supplementary Figure S19). However, Bh. rubra BCC47515 did not produce these molecules in our study, suggesting it may be a strain specific ability. The lack of bioactivity may come as a surprise, given the fact that previously reported penicolinates A-E exhibited antibacterial, antifungal and antiplasmodal activity (Intaraudom et al. 2013). Due to limited amount of material, penicolinates F and G could not be tested against plasmodia and an acitivity cannot be excluded. Having a free alkyl chain, penicolinate G (2) may also be a promising candidate in a biofilm inhibition assay, as such structural moieties have shown to be favorable in inhibiting biofilms (Becker et al. 2020; Chepkirui et al. 2018).
Data availability
All sequence data generated in this study (see Table 1) are available in GenBank (https://www.ncbi.nlm.nih.gov/genbank/).
References
Alali S, Mereghetti V, Faoro F, Bocchi S, Al Azmeh F, Montagna M (2019) Thermotolerant isolates of Beauveria bassiana as potential control agent of insect pest in subtropical climates. PloS One 14:e0211457. https://doi.org/10.1371/journal.pone.0211457
Alagesan A, Tharani G, Padmanaban B, Manivannan S, Jawahar S (2019) An assessment of biological control of the banana pseudostem weevil Odoiporus longicollis (Olivier) by entomopathogenic fungi Beauveria bassiana. Biocatal Agric Biotechnol 20:101262. https://doi.org/10.1016/j.bcab.2019.101262
Araújo JPM, Lebert BM, Vermeulen S, Brachmann A, Ohm RA, Evans HC, Debekker C (2022) Masters of the manipulator: two new hypocrealean genera, Niveomyces (Cordycipitaceae) and Torrubiellomyces (Ophiocordycipitaceae), parasitic on the zombie ant fungus Ophiocordyceps camponoti-floridani. Persoonia 49:171–194. https://doi.org/10.3767/persoonia.2022.49.05
Avery PB, Faull J, Simmonds MSJ (2004) Effect of different photoperiods on the growth, infectivity and colonization of Trinidadian strains of Paecilomyces fumosoroseus on the greenhouse whitefly, Trialeurodes vaporariorum, using a glass slide bioassay. J Insect Sci 4:38. https://doi.org/10.1093/jis/4.1.38
Avery PB, Faull J, Simmonds MSJ (2008) Effects of Paecilomyces fumosoroseus and Encarsia formosa on the control of the greenhouse whitefly: Preliminary assessment of a compatibility study. Biocontrol 53:303–316. https://doi.org/10.1007/s10526-007-9073-5
Aynalem B, Muleta D, Jida M, Shemekite F, Aseffa F (2022) Biocontrol competence of Beauveria bassiana, Metarhizium anisopliae and Bacillus thuringiensis against tomato leaf miner, Tuta absoluta Meyrick 1917 under greenhouse and field conditions. Heliyon 8:e09694. https://doi.org/10.1016/j.heliyon.2022.e09694
Barra-Bucarei L, González MG, Iglesias AF, Aguayo GS, Peñalosa MG, Vera PV (2020) Beauveria bassiana multifunction as an endophyte: growth promotion and biologic control of Trialeurodes vaporariorum (Westwood) (Hemiptera: Aleyrodidae) in Tomato. Insects 11:591. https://doi.org/10.3390/insects11090591
Becker K, Pfütze S, Kuhnert E, Cox RJ, Stadler M, Surup F (2020) Hybridorubrins A-D: Azaphilone heterodimers from stromata of Hypoxylon fragiforme and insights into the biosynthetic machinery for azaphilone diversification. Chemistry Eur J 27(4):1. https://doi.org/10.1002/chem.202003215
Bischoff JF, White JF Jr (2004) Torrubiella piperis sp. nov. (Clavicipitaceae, Hypocreales), a new teleomorph of the Lecanicillium complex. Stud Mycol 50:89–94
Bischoff JF, Chaverri P, White JF Jr (2005) Clarification of the host substrate of Ascopolyporus and description of Ascopolyporus philodendrus sp. nov. Mycologia 97:710–717. https://doi.org/10.1080/15572536.2006.11832800
Bosch A, Yantorno O (1999) Microcycle conidiation in the entomopathogenic fungus Beauveria bassiana Bals. (Vuill.). Process Biochem 34:707–716. https://doi.org/10.1016/S0032-9592(98)00145-9
Chaverri P, Bischoff J, Evans H, Hodge K (2005) Regiocrella, a new entomopathogenic genus with a pycnidial anamorph and its phylogenetic placement in the Clavicipitaceae. Mycologia 97:1225–1237
Chen W, Liang J, Ren X, Zhao J, Han Y, Liang Z (2022a) Multigene phylogeny, phylogenetic network, and morphological characterizations reveal four new arthropod-associated Simplicillium species and their evolutional relationship. Front Microbiol 4(13):950773. https://doi.org/10.3389/fmicb.2022.950773
Chen WH, Han YF, Liang JD, Tian WY, Liang ZQ (2021a) Multi-gene phylogenetic evidence indicates that Pleurodesmospora belongs in Cordycipitaceae (Hypocreales, Hypocreomycetidae) and Pleurodesmospora lepidopterorum sp. nov. on pupa from China. MycoKeys 80:45–55. https://doi.org/10.3897/mycokeys.80.66794
Chen WH, Liu C, Han YF, Liang JD, Liang ZQ (2018) Akanthomyces araneogenum, a new isaria-like araneogenous species. Phytotaxa 379:66–72. https://doi.org/10.11646/phytotaxa.379.1.6
Chen WH, Han YF, Liang JD, Tian WY, Liang ZQ (2020) Morphological and phylogenetic characterisations reveal three new species of Samsoniella (Cordycipitaceae, Hypocreales) from Guizhou, China. MycoKeys 74:1–15. https://doi.org/10.3897/mycokeys.74.56655
Chen W, Liang J, Ren X, Zhao J, Han Y, Liang Z (2021b) Cryptic diversity of isaria-like species in Guizhou. China Life 11:1093. https://doi.org/10.3390/life11101093
Chen WH, Liu C, Han YF, Liang JD, Tian WY, Liang ZQ (2019) Akanthomyces araneicola, a new araneogenous species from Southwest China. Phytotaxa 409:227–232. https://doi.org/10.11646/phytotaxa.409.4.5
Chen WH, Liu C, Liang JD, Ren XX, Zhao JH, Han YF (2022b) Species diversity of cordyceps-like fungi in the Tiankeng Karst Region of China. Microbiol Spectr 10:e0197522. https://doi.org/10.1128/spectrum.01975-22
Chepkirui C, Yuyama KT, Wanga LA, Decock C, Matasyoh JC, Abraham WR, Stadler M (2018) Microporenic acids A-G, biofilm Inhibitors, and antimicrobial agents from the basidiomycete Microporus species. J Nat Prod 81:778–784. https://doi.org/10.1021/acs.jnatprod.7b00764
Chiriví J, Danies G, Sierra R, Schauer N, Trenkamp S, Restrepo S, Sanjuan T (2017) Metabolomic profile and nucleoside composition of Cordyceps nidus sp. nov. (Cordycipitaceae): a new source of active compounds. PLoS One 12:e0179428. https://doi.org/10.1371/journal.pone.0179428
Crous PW, Wingfield MJ, Lombard L et al (2019) Fungal Planet description sheets: 951–1041. Persoonia 43:223–425. https://doi.org/10.3767/persoonia.2019.43.06
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Helaly SE, Kuephadungphan W, Phainuphong P, Ibrahim MAA, Tasanathai K, Mongkolsamrit S, Luangsa-ard JJ, Phongpaichi S, Rukachaisirikul V, Stadler M (2019) Pigmentosins from Gibellula sp. as anti-biofilm agents and a new glycosylated asperfuran derivative from Cordyceps javanica. Beilstein J Org Chem 15:2968–2981. https://doi.org/10.3762/bjoc.15.293
Hall T (1999) BioEdit. a user-friendly biological sequence alignment editor and analysis program forWindows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Intaraudom C, Boonyuen N, Suvannakad R, Rachtawee P, Pittayakhajonwut P (2013) Penicolinates A-E from endophytic Penicillium sp. BCC16054. Tetrahedron Lett 54:744–748
Jędrejko KJ, Lazur J, Muszyńska B (2021) Cordyceps militaris: An overview of its chemical constituents in relation to biological activity. Foods 10:2634. https://doi.org/10.3390/foods10112634
Johnson D, Sung GH, Hywel-Jones NL, Luangsa-Ard JJ, Bischoff JF, Kepler RM, Spatafora JW (2009) Systematics and evolution of the genus Torrubiella (Hypocreales, Ascomycota). Mycol Res 113:279–289. https://doi.org/10.1016/j.mycres.2008.09.008
Keçili S, Bakır A, Kutalmış A, Çelik T, Sevim A (2022) Soil isolation, identification, and virulence testing of Turkish entomopathogenic fungal strains: a potential native isolate of Beauveria bassiana for the control of Leptinotarsa decemlineata. Biocontrol 67:593–603. https://doi.org/10.1007/s10526-022-10156-4
Kepler RM, Luangsa-ard JJ, Hywel-Jones NL, Quandt CA, Sung GH, Rehner SA, Aime MC, Henkel TW, Sanjuan T, Zare R, Chen M, Li Z, Rossman AY, Spatafora JW, Shrestha B (2017) A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8:335–353. https://doi.org/10.5598/imafungus.2017.08.02.08
Kepler RM, Sung GH, Ban S, Nakagiri A, Chen MJ, Huang B, Li Z, Spatafora JW (2012) New teleomorph combinations in the entomopathogenic genus Metacordyceps. Mycologia 104:182–197. https://doi.org/10.3852/11-070
Khonsanit A, Luangsa-ard JJ, Thanakitpipattana D, Noisripoom W, Chaitika T, Kobmoo N (2020) Cryptic diversity of the genus Beauveria with a new species from Thailand. Mycol Prog 19:291–315. https://doi.org/10.1007/s11557-020-01557-9
Kuephadungphan W, Macabeo APG, Luangsa-ard JJ, Tasanathai K, Thanakitpipattana D, Phongpaichit S, Yuyama K, Stadler M (2019) Studies on the biologically active secondary metabolites of the new spider parasitic fungus Gibellula gamsii. Mycol Prog 18:135–146. https://doi.org/10.1007/s11557-018-1431-4
Kuephadungphan W, Tasanathai K, Petcharad B, Khonsanit A, Stadler M, Luangsaard JJ (2020) Phylogeny- and morphology-based recognition of new species in the spider-parasitic genus Gibellula (Hypocreales, Cordycipitaceae) from Thailand. MycoKeys 72:17–42. https://doi.org/10.3897/mycokeys.72.55088
Kuephadungphan W, Pritchard B, Tasanathai K, Thanakitpipattana D, Kobmoo N, Khonsanit A, Samson RA, Luangsaard JJ (2022) Multi-locus phylogeny unmasks hidden species within the specialised spider parasitic fungus, Gibellula (Hypocreales, Cordycipitaceae). Stud Mycol 101:245–286. https://doi.org/10.3114/sim.2022.101.04
Luangsa-Ard JJ, Hywel-Jones NL, Manoch L, Samson RA (2005) On the relationships of Paecilomyces sect. Isarioidea species. Mycol Res 109:581–589. https://doi.org/10.1017/S0953756205002741
Mendes-Pereira T, de Araújo JPM, Kloss TG, Costa-Rezende DH, de Carvalho DS, Góes-Neto A (2023) Disentangling the taxonomy, systematics, and life history of the spider-parasitic fungus Gibellula (Cordycipitaceae, Hypocreales). J Fungi 9:457. https://doi.org/10.3390/jof9040457
Mongkolsamrit S, Noisripoom W, Hasin S, Phirada S, Panrada J, Luangsa-ard JJ (2023) Multi-gene phylogeny and morphology of Ophiocordyceps laotii sp. nov. and a new record of O. buquetii (Ophiocordycipitaceae, Hypocreales) on ants from Thailand. Mycol Prog 22:5. https://doi.org/10.1007/s11557-022-01855-4
Mongkolsamrit S, Noisripoom W, Pumiputikul S, Boonlarppradab C, Samson RA, Stadler M, Becker K, Luangsa-ard JJ (2021) Ophiocordyceps flavida sp. nov. (Ophiocordycipitaceae), a new species from Thailand associated with Pseudogibellula formicarum (Cordycipitaceae), and their bioactive secondary metabolites. Mycol Prog 20:477–492. https://doi.org/10.1007/s11557-021-01683-y
Mongkolsamrit S, Noisripoom W, Tasanathai K, Khonsanit A, Thanakitpipattana D, Himaman W, Kobmoo N, Luangsa-ard JJ (2020) Molecular phylogeny and morphology reveal cryptic species in Blackwellomyces and Cordyceps (Cordycipitaceae) from Thailand. Mycol Prog 19:957–983. https://doi.org/10.1007/s11557-020-01615-2
Mongkolsamrit S, Noisripoom W, Tasanathai K, Kobmoo N, Thanakitpipattana D, Khonsanit A, Petcharad B, Sakolrak B, Himaman W (2022) Comprehensive treatise of Hevansia and three new genera Jenniferia, Parahevansia and Polystromomyces on spiders in Cordycipitaceae from Thailand. MycoKeys 91:113–149. https://doi.org/10.3897/mycokeys.91.83091
Mongkolsamrit S, Noisripoom W, Thanakitpipattana D, Wutikhun T, Spatafora JW, Luangsa-ard J (2018) Disentangling cryptic species with isaria-like morphs in Cordycipitaceae. Mycologia 110:230–257. https://doi.org/10.1080/00275514.2018.1446651
Nishi O, Sushida H, Higashi Y, Iida Y (2021) Entomopathogenic fungus Akanthomyces muscarius (Hypocreales: Cordycipitaceae) strain IMI268317 colonises on tomato leaf surface through conidial adhesion and general and microcycle conidiation. Mycology 13:133–142. https://doi.org/10.1080/21501203.2021.1944929
Nylander JAA (2004) MrModeltest Version 2. Evolutionary Biology Centre, Uppsala University, Uppsala, Program distributed by the author
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358. https://doi.org/10.1093/bioinformatics/12.4.357
Perdomo H, Cano J, Gené J, García D, Hernández M, Guarro J (2013) Polyphasic analysis of Purpureocillium lilacinum isolates from different origins and proposal of the new species Purpureocillium lavendulum. Mycologia 105:151–161. https://doi.org/10.3852/11-190
Ramakuwela T, Hatting J, Bock C, Vega FE, Wells L, Mbata GN, Shapiro-Ilan D (2020) Establishment of Beauveria bassiana as a fungal endophyte in pecan (Carya illinoinensis) seedlings and its virulence against pecan insect pests. Biol Control 140:104102. https://doi.org/10.1016/j.biocontrol.2019.104102
Rehner SA, Minnis AM, Sung GH, Luangsa-ard JJ, Devotto L, Humber RA (2011) Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia 103:1055–1073. https://doi.org/10.3852/10-302
Ronquist F, Teslenko M, van der Mark P, Ayres DL, DarlingA HS, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference andmodel choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Royal Horticultural Society Colour Chart (RHS Colour Chart), 6th ed.; Royal Horticultural Society: London, UK, 2015
Sandargo B, Kaysan L, Teponno RB, Richter C, Thongbai B, Surup F, Marc S (2021) Analogs of the carotane antibiotic fulvoferruginin from submerged cultures of a Thai Marasmius sp. Beilstein J Org Chem 17:1385–1391. https://doi.org/10.3762/bjoc.17.97
Sanjuan T, Tabima J, Restrepo S, Læssøe T, Spatafora J, Molano A (2014) Entomopathogens of Amazonian stick insects and locusts are members of the Beauveria species complex (Cordyceps sensu stricto). Mycologia 106:260–275. https://doi.org/10.3852/13-020
Schrey H, Scheele T, Ulonska C, Nedder DL, Neudecker T, Spiteller P, Stadler M (2022) Alliacane-type secondary metabolites from submerged cultures of the basidiomycete Clitocybe nebularis. J Nat Prod 85:2363–2371. https://doi.org/10.1021/acs.jnatprod.2c00554
Shrestha B, Kubátová A, Tanaka E, Oh J, Yoon DH, Sung JM, Sung GH (2019) Spider-pathogenic fungi within Hypocreales (Ascomycota): Their current nomenclature, diversity, and distribution. Mycol Prog 18:983–1003. https://doi.org/10.1007/s11557-019-01512-3
Shrestha B, Tanaka E, Hyun MW, Han JG, Kim CS, Jo JW, Han SK, Oh J, Sung GH (2016) Coleopteran and lepidopteran hosts of the entomopathogenic genus Cordyceps sensu lato. J Mycol 7648219:1–14. https://doi.org/10.1155/2016/7648219
Spatafora JW, Sung GH, Sung JM, Hywel-Jones NL, White JF Jr (2007) Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol Ecol 16:1701–1711. https://doi.org/10.1111/j.1365-294X.2007.03225.x
Song D, Shi Y, Ji H, Xia Y, Peng G (2019) The MaCreA Gene Regulates Normal conidiation and microcycle conidiation in Metarhizium acridum. Front Microbiol 10:1946. https://doi.org/10.3389/fmicb.2019.01946
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Sung GH, Hywel-Jones NL, Sung JM, Luangsa-ard JJ, Shrestha B, Spatafora JW (2007) Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol 57:5–59. https://doi.org/10.3114/sim.2007.57.01
Sung GH, Spatafora JW (2004) Cordyceps cardinalis sp. nov., a new species of Cordyceps with an east Asian-eastern North American distribution. Mycologia 96:658–666. https://doi.org/10.1080/15572536.2005.11832962
Sung GH, Spatafora JW, Zare R, Hodge KT, Gams W (2001) A revision of Verticillium sect. Prostrata. II. Phylogenetic analyses of SSU and LSU nuclear rDNA sequences from anamorphs and teleomorphs of the Clavicipitaceae. Nova Hedw 72:311–328
Tsang CC, Chan JFW, Pong WM, Chen JHK, Ngan AHY, Cheung M, Lai CKC, Tsang DNC, Lau SKP, Woo PCY (2016) Cutaneous hyalohyphomycosis due to Parengyodontium album gen. et comb. nov. Med Mycol 54:699–713. https://doi.org/10.1093/mmy/myw025
Thanakitpipattana D, Tasanathai K, Mongkolsamrit S, Khonsanit A, Lamlertthon S, Luangsa-ard J (2020) Fungal pathogens occurring on Orthopterida in Thailand. Persoonia 44:140–160. https://doi.org/10.3767/persoonia.2020.44.0600457-3
Thanakitpipattana D, Mongkolsamrit S, Khonsanit A, Himaman W, Luangsa-ard JJ, Pornputtapong N (2022) Is Hyperdermium congeneric with Ascopolyporus? Phylogenetic relationships of Ascopolyporus spp (Cordycipitaceae, Hypocreales) and a new genus Neohyperdermium on scale insects in Thailand. J Fungi 8:516. https://doi.org/10.3390/jof8050516
Vu D, Groenewald M, de Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T, Crous PW, Robert V, Verkley GJM (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom Fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud Mycol 92:135–154. https://doi.org/10.1016/j.simyco.2018.05.001
Wang YB, Wang Y, Fan Q, Duan DE, Zhang GD, Dai RQ, Dai YD, Zeng WB, Chen ZH, Li DD, Tang DX, Xu ZH, Sun T, Nguyen TT, Tran NL, Dao VM, Zhang CM, Huang LD, Liu YJ, Zhang XM, Yang DR, Sanjuan T, Liu XZ, Yang ZL, Yu H (2020) Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers 103:1–46. https://doi.org/10.1007/s13225-020-00457-3
Wang Z, Wang Y, Dong Q, Fan Q, Dao V-M, Yu H (2022) Morphological and phylogenetic characterization reveals five new species of Samsoniella (Cordycipitaceae, Hypocreales). J Fungi 8:747. https://doi.org/10.3390/jof8070747
Wang Z, Jin K, Xia Y (2016) Transcriptional analysis of the conidiation pattern shift of the entomopathogenic fungus Metarhizium acridum in response to different nutrients. BMC Genomics 17:586. https://doi.org/10.1186/s12864-016-2971-0
Wei DP, Wanasinghe DN, Hyde KD, Mortimer PE, Xu J, Xiao YP, Bhunjun CS, To-anun C (2019) The Genus Simplicillium. Mycokeys 60:69–92. https://doi.org/10.3897/mycokeys.60.38040
Zare R, Gams W (2001) A revision of Verticillium section Prostrata. IV. The genera Lecanicillium and Simplicillium gen. nov. Nova Hedw 73:1–50
Zare R, Gams W (2008) A revision of the Verticillium fungicola species complex and its affinity with the genus Lecanicillium. Mycol Res 112:811–824. https://doi.org/10.1016/j.mycres.2008.01.019
Zhang X, Hu Q, Weng Q (2018) Secondary metabolites (SMs) of Isaria cicadae and Isaria tenuipes. RSC Adv 9(1):172–184. https://doi.org/10.1039/c8ra09039d
Zimmermann G (2008) The entomopathogenic fungi Isaria farinosa (formerly Paecilomyces farinosus) and the Isaria fumosorosea species complex (formerly Paecilomyces fumosoroseus): Biology, ecology and use in biological control. Biocontrol Sci Technol 18:865–901
Zhang L, Fasoyin OE, Molnár I, Xu Y (2020) Secondary metabolites from hypocrealean entomopathogenic fungi: Novel bioactive compounds. Nat Prod Rep 37:1181–1206. https://doi.org/10.1039/c9np00065h
Zhang ZF, Zhou SY, Eurwilaichitr L, Ingsriswang S, Raza M, Chen Q, Zhao P, Liu F, Cai L (2021) Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Divers 106:29–136. https://doi.org/10.1007/s13225-020-00453-7
Zhou YM, Zhi JR, Qu JJ, Zou X (2022) Estimated divergence times of Lecanicillium in the family Cordycipitaceae provide insights into the attribution of Lecanicillium. Front Microbiol 13:859886. https://doi.org/10.3389/fmicb.2022.859886
Zou Y, Li C, Wang S, Xia Y, Jin K (2022) MaCts1 an endochitinase, Is involved in conidial germination, conidial yield, stress tolerances and microcycle conidiation in Metarhizium acridum. Biology 11:1730. https://doi.org/10.3390/biology11121730
Acknowledgements
We are indebted to the Department of National Parks, Wildlife and Plant Conservation for their cooperation and support of our research project. The authors would like to thank the former colleagues Wilawan Kuephadungphan and Soleiman E. Helaly for initiating the crude extraction and purification of fungal metabolites. Wera Collisi and Christel Kakoschke are acknowledged for their excellent technical support.
Funding
Open Access funding enabled and organized by Projekt DEAL. This research benefitted from funding by the European Union’s Horizon 2020 research and innovation program (RISE) under the Marie Skłodowska-Curie grant agreement No. 101008129, project acronym “MycoBiomics” (lead beneficiaries J.J.L. and M.S.). We are also grateful to the the National Science and Technology Development Agency (NSTDA for a grant to the National Center for Genetic Engineering and Biotechnology (BIOTEC) Platform Technology Management (no. P19-50231). S.S.E. gratefully acknowledges Alexander von Humboldt (AvH) Foundation for funding him via a Georg-Forster Fellowship for Experienced Researchers (Ref 3.4–1222288-EGY-GF-E).
Author information
Authors and Affiliations
Contributions
S.M., B.S., W.N., S.J.: investigation, writing—original draft preparation; B.S., S.S.E.: compound isolation, structure elucidation; M.S., J.J. L.: conceptualization, supervision, writing – original draft preparation, resources, review and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Additional information
Section Editor: Ji-Kai Liu
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mongkolsamrit, S., Sandargo, B., Ebada, S.S. et al. Bhushaniella gen. nov. (Cordycipitaceae) on spider eggs sac: a new genus from Thailand and its bioactive secondary metabolites. Mycol Progress 22, 64 (2023). https://doi.org/10.1007/s11557-023-01915-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-023-01915-3