Abstract
Tropical dry forests are an intricate ecosystem with special adaptations to periods of drought. Arbuscular mycorrhizal fungi (AMF) are essential for plant survival in all terrestrial ecosystems but might be of even greater importance in dry forests as plant growth is limited due to nutrient and water deficiency during the dry season. Tropical dry forests in Ecuador are highly endangered, but studies about AMF communities are scarce. We investigated the AMF community of a premontane semi-deciduous dry forest in South Ecuador during the dry season. We estimated AMF diversity, distribution, and composition of the study site based on operational taxonomic units (OTUs) and compared the results to those from the tropical montane rainforest and páramo in South Ecuador. OTU delimitation was based on part of the small ribosomal subunit obtained by cloning and Sanger sequencing. Nearly all OTUs were Glomeraceae. The four frequent OTUs were Glomus, and comparison with the MaarjAM database revealed these to be globally distributed with a wide range of ecological adaptations. Several OTUs are shared with virtual taxa from dry forests in Africa. Ordination analysis of AMF communities from the tropical dry and montane rainforests in South Ecuador revealed a unique AMF community in the dry forest with only few overlapping OTUs. Most OTUs that were found in both dry and rainforests and on the two continents were globally distributed Glomus.
Similar content being viewed by others
Introduction
Tropical dry forests are a unique biome that can be found in the tropics of South America, Central America, Africa, and Eurasia (Miles et al. 2006). These forests receive less than ~1800 mm rainfall per year and have a dry period from 3 up to 6 months during which most of the vegetation drop their leaves. Tropical dry forests are diverse ecosystems with high levels of endemism and floristic turnover (DRYFLOR et al. 2016). Only 10% of their original extent remains today in many countries (DRYFLOR et al. 2016), and those remnants are highly endangered due to climate change, habitat fragmentation, and conversion into farmland (Miles et al. 2006).
The Neotropics contain most of all remaining tropical dry forest on Earth with more than half in South America alone (Miles et al. 2006). Neotropical dry forests are distinguished into 12 groups, based on the occurrence and frequency of their woody species (DRYFLOR et al. 2016): (i) Mexico, (ii) Antilles, (iii) Central America–northern South America, (iv) northern inter-Andean valleys, (v) central inter-Andean valleys, (vi) central Andes coast, (vii) Tarapoto-Quillabamba, (viii) Apurimac-Mantaro, (ix) Piedmont, (x) Misiones, (xi) central Brazil, and (xii) Caatinga. For most of the floristic groups, no studies about mycorrhizal communities exist, with the exception of Mexico (i) (Aguilar-Fernandéz et al. 2009, Carillo-Saucedo et al. 2018, Guadarrama et al. 2014) and Caatinga (xii) in Brazil (Sousa et al. 2018, Marinho et al. 2019, Teixeira-Rios et al. 2018). All of these studies used spores to identify AMF, and based on that, the results show a high level of agreement with dominant representatives of the Glomeraceae and Acaulosporaceae. A better understanding of AMF communities is of utter importance to understand ecosystem dynamics and dependencies and to protect the remaining forests (Lugo & Pagano 2019).
We present the first results of AMF community composition for one of the two floristic groups in Ecuador—the Central Andes Coast Forests (vi). To our knowledge, this is the first study about AMF in neotropical dry forests in Ecuador.
The study site is a premontane semi-deciduous dry forest in the Reserva National Laipuna. This forest, also called ceibo forest, is famous for its ceiba trees (Ceiba trichistandra), which characterize large areas of the provinces of Manabí, Guayas, El Oro, and Loja (Aguirre et al. 2006). Tabebuia chrysantha and species of Bombacaceae are dominating floristic elements in these forests (Aguirre et al. 2006).
The aim of our investigation was to gain knowledge about AMF community composition analyzing root and soil samples with molecular methods and to compare the results to our extensive findings from neotropical rain forests in Southern Ecuador (Haug et al. 2019).
Material and methods
Study area
The study was conducted in the “Reserva National Laipuna” (4° 22′ S–79° 90′ W) in Southern Ecuador, which belongs to the Tumbesian dry forest ecoregion (Fig. S1). This area is located about 200 km west from our lower and upper rain forest sample sites (for details see Haug et al. 2019). Laipuna is characterized by hilly terrain and altitudes of 600–1450 masl. Precipitation strongly varies from year to year but ranges between 350 and 800 mm (Pucha-Cofrep et al. 2015). Annual precipitation distribution shows a distinctive dry season from June to December and a wet season from January to May. Annual mean temperature is 23.4 °C and shows little variability throughout the year (Pucha-Cofrep et al. 2015).
Vegetation
The ecosystem is classified as a premontane semi-deciduous dry forest (Aguirre et al. 2006). Frequent tree species are Tabebuia chrysantha, Eriotheca ruizii, Ceiba trichistandra, Erythrina velutina, Ipomoea wolcottiana, Bursera graveolens, Terminalia valverdae, and Fulcadea laurifolia (J. Homeier, unpublished).
Sample collection
Soil and mycorrhizal roots from the topsoil (2–10 cm depth) were sampled during the dry season in October 2016. The organic layer was loosened with a rake, and sections of several fine-root systems were sampled in plastic bags. Each fine-root system was placed in a separate PCR tube to ensure that each mycorrhizal sample is from a single host plant. Mycorrhizas and soil samples were dried at about 50 °C for 24 h by placing open sample tubes on an electric dryer. After the drying step, silica gel was added, and the tubes were closed for long-term storage.
In total, we collected 95 root and 38 soil samples across sites between 600 and 1250 masl.
DNA isolation
Approximately 5–10 root segments with a length of 2 cm or about 50 mg soil were ground using a carbide ball and a mixer mill (2×1.30 min, frequency 30/s). Total DNA was isolated with either the innuPREP Plant DNA Kit (Analytik Jena, Germany) or the DNeasy Power Soil Kit (Qiagen, Germany) and re-suspended in a final volume of 100 μl elution buffer.
PCR, cloning, and Sanger sequencing
Part of the SSU region of the nuclear ribosomal rDNA repeat was amplified by PCR using a volume of 0.5 μl of the DNA template. A nested PCR approach was applied, amplifying the larger outer fragment with the primer combination NS1/NS4 (White et al. 1990, Lee et al. 2008) and the smaller, inner fragment with the primer pair AML1/AML2 (Lee et al. 2008; for details see Haug et al. 2013). We chose the SSU as the target region because it has been widely used (Öpik et al. 2013, Van Geel et al. 2014); this primer pair is suitable to detect all AMF subgroups (Lee et al. 2008, Van Geel et al. 2014) and provides good comparability with previous studies from other regions (e.g., MaarjAM database: Öpik et al. 2010). It is important to note that AMF species diversity is highly impacted by DNA region, similarity thresholds, and cluster algorithms (Göker et al. 2010), but our main focus is on comparison of AMF composition among different sites and diversity assessment of dry forest AMF.
Amplified PCR products were cloned with the Invitrogen TA Cloning Kit (Life Technologies) following the manufacturer’s instructions but using a third of the indicated volumes. Inserts were re-amplified from clones with primers M13F/M13R by picking eight bacterial clones with a separate toothpick and stirring them into one PCR reaction mix per clone. Inserts were digested with restriction enzymes HinfI or AfaI. Digested products were examined on a 0.7% agarose gel. Two clones of the same RFLP pattern were cleaned and sequenced by GATC Biotech (Konstanz, Germany). Whenever the two sequences were not identical, two additional clones of the same RFLP pattern were sequenced. Non-Glomeromycotina sequences (~5% of inserts) were disregarded.
Glomeromycotina sequences were edited with Sequencher (Version 4.9, Gene Codes, Ann Arbor, Michigan), and a BLAST search was performed against the nucleotide sequence database (NCBI) and against the MaarjAM database. When several inserts of a cloned PCR product belonged to the same OTU, only one sequence was included in the final data set.
The final data set (Table S1) consisted of 215 Glomeromycotina sequences: 152 sequences generated from mycorrhizal samples and 63 sequences from soil samples. The sequences were deposited in GenBank (SAMN14381484–SAMN14381698, Table S1).
OTU delimitation
All sequences were ~800 bp long and comprised part of the 18S. Sequences were aligned with MAFFT version 7 (https://mafft.cbrc.jp/alignment/server/ (Katoh et al. 2005); MAFFT-L-INS-i), and a distance matrix based on p-distances was created with PAUP for OTU delimitation.
Subsequently, operational taxonomic units (OTUs) were defined as surrogates for species based on sequence similarity with OPTSIL (Göker et al. 2010). We used intermediate linkage clustering and a cut-off value of 99% sequence similarity. This approach splits OTUs if 50% or more of their containing sequences are less than 99% similar to another OTU (for details see Göker et al. 2010). This is the same OTU delimitation approach as in Kottke et al. (2008), Haug et al. (2013), and Haug et al. (2019) to allow a comprehensive comparison with OTUs from the rainforest of Southern Ecuador.
For better comparability, the OTU numbers of the rainforest from the 2019 study (Haug et al. 2019) were retained and marked with an R. The new dry forest OTUs were prefixed with the letter “D” and numbered from 1 to 16.
Singleton sequences (10 in total) were excluded from subsequent analyses.
AMF community analyses
We combined all 152 sequences from mycorrhizal dry forest samples with our existing data set from the mycorrhizal rain forest samples (Haug et al. 2019).
To explore differences in AMF community compositions among sampling sites, we carried out a non-metric multidimensional scaling (NMDS) ordination with metaMDS from the R package VEGAN (Oksanen et al. 2018). The underlying matrix was a Kulczynski dissimilarity matrix derived from OTU abundances of each site. The Kulczynski dissimilarity measure was given the highest rank using R’s rankindex test in VEGAN. The metaMDS function in VEGAN allows for calculating the NMDS ordination multiple times with random starts in order to find a stable result (https://www.rdocumentation.org/packages/vegan/versions/2.4-2/topics/metaMDS). We used a maximum number of 1000 iterations in a 3-dimensional space.
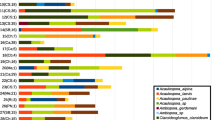
OTU abundance per forest site. Relative abundance of OTUs occurring in > 10% of samples in at least one forest site. The dry forest (orange) and rain forest at 1000 m (green) show an AMF community that is dominated by few frequent OTUs (e.g., OTUR31 from the dry forest and OTUR17 from the 1000 m rain forest occurred in 39% of all samples each) not found at most other sites. Abundant OTUs from higher elevation forests (≥ 2000 m) show a wide distribution across different altitudes, but even the most frequent OTUs occur only in 16% of all samples
We performed pairwise PERMANOVA (pairwise.adonis, R) to test for OTU similarity between each site using Holm’s correction to control for multiple comparisons.
Results and discussion
AMF community composition of the tropical dry forest in Laipuna
The first results for AMF in a tropical dry forest in Ecuador revealed 27 OTUs from 215 sequences and 133 samples. The rarefaction curve nearly reached the asymptote (Fig. S2) suggesting that few more OTUs are to be found.
Almost all OTUs (93%) belonged to the genus Glomus (Figs. 1 and 2) and were found in mycorrhizal samples as well as in the soil (Fig. 3, Table 1). The most abundant OTU was Glomus OTU-R31 (= VT69) occurring in 39% of all samples (Fig. 4, Table 1) followed by Glomus OTU-R57 (= VT166) found in 23% of the samples. Further frequent OTUs were Glomus OTU-D7 (= VT155) in 13% and OTU-R7 (= VT248) in 11% of the samples (Fig. 4, Table 1). All of these four Glomus OTUs are globally distributed, occurring in a variety of biomes (MaarjAM database, https://maarjam.botany.ut.ee/). We therefore assume that the dominating Glomus OTUs in Laipuna have a range of ecological adaptations and are not specific to dry forest habitats.
In addition to the four dominant Glomus OTUs, there were many rare Glomus OTUs (Fig. 5, Table 1). Besides the genus Glomus, only two more genera, Claroideoglomus and Diversispora, were found but both in very low numbers. Diversispora was represented by only a single sequence (1 OTU) and Claroideoglomus by one OTU and four sequences (Figs. 1 and 2, Table 1). The Claroideoglomus OTU has a worldwide distribution (VT193 – MaarjAM), and the Diversispora OTU has a close match (98.5% similarity) to VT262 from the MaarjAM database that also occurs in French Polynesia and Micronesia.
The lack of AMF groups other than Glomus was unexpected as studies from dry forests in Brazil, yet spore-based and not molecular, showed frequent occurrence of Acaulosporaceae, Gigasporales, and Glomerales (Pagano et al. 2019).
Comparison of AMF from Laipuna with AMF in tropical rain forests of Southern Ecuador
We compared all Laipuna OTUs isolated from mycorrhizal samples at 600–1250 masl with AMF from tropical rain forest sites in Southern Ecuador at 1000–4000 masl (Fig. 6) (Haug et al. 2019). The ordination results show a separate AMF community for the dry forest in Laipuna (Fig. 7). This is supported by the results of pairwise PERMANOVA showing significant differences between the dry forest and the rainforest for all elevation sites above 1000 masl (Table 2). There is some OTU overlap between the dry forest and the lower altitudes of the rainforest (1000 and 2000 masl). Despite the large discrepancies in cloud frequency (Beck et al. 2008), annual rainfall (350–800 mm versus 1950–2230 mm/year), and tree species composition, the AMF community in Laipuna showed similarities with the 1000 masl level by having dominating Glomus members and an overlap of seven OTUs (Figs. 4 and 6, Table 1). These overlaps are in agreement with the results of Davison et al. (2021) which found that temperature is one of the most important abiotic drivers for AMF distribution. A distinct AMF community in the dry forest does not necessarily suggest that dry forests AMF are also dry forest specialists as community patterns can be the result of species distribution instead of habitat adaptation (Gai et al. 2006). From studies of AMF in tropical dry and rainforests (Marinho et al. 2018), we know that many species were found in both, dry, and rain forest sites. In addition, many AM fungi seem to be adaptable to different water regimes and thrive in different biomes and ecosystems (Davison et al. 2015). Whether the initial results we are presenting here hold up with more rigorous sampling remains to be seen. The rarefaction curve (Fig. S2) shows that sampling was not yet exhaustive. Also, we suggest that in future studies, samples from the wet season should be included as much as possible as frequencies of OTUs, and occurrences might shift between seasons. Differences in AMF species abundance between dry and wet seasons, but no difference in species richness was reported in da Silva et al. (2014) and Maitra et al. (2019). Guadarrama et al. (2014) found differences between the dry and wet season but only for three species—all of them non-Glomus AMF: Gigaspora gigantea only in the dry season and Acaulospora foevata and Scutellospora erythropus exclusively in the wet season. On the contrary, Teixeira-Rios et al. (2018) found no statistical difference for AMF communities during dry and wet seasons.
Shared and unique OTUs of Laipuna Dry Forest with respect to elevation belts of the rain forest. The horizontal bars connecting the same sites indicate the amount of unique OTUs for this site. The thickness of connector lines between different sites represents the percentage of OTUs shared. Laipuna had a high number of unique OTUs and shared several OTUs with the 1000 and 2000 m sites
a NMDS ordination shows turnover of species among sites. Species scores are shown as gray dots and site scores as black labels (see Table S2 for more info). The ellipses around data points show the 95% confidence interval for Laipuna dry forest (pink), the evergreen premontane forest (1000 m, orange), evergreen lower montane forest (2000 m, dark green), upper montane forest (3000 m, light green), and grass paramo/Polylepis forest (4000 m, blue) sites. The confidence interval for Laipuna is not overlapping with any of the rainforest sites, indicating a distinct AMF community. This is supported by PERMANOVA for site comparison Laipuna and upper montane forest (F= 14.285, p= 0.033), Laipuna and grass paramo/Polylepis (F= 8.937, p= 0.041), and Laipuna and evergreen lower montane forest (F= 8.706, p= 0.041). Overall stress of the ordination is 0.073 with 3 dimensions (k= 3). b Shepard diagram of the NMDS ordination showing the relationship of the observed dissimilarity with the ordination distance calculated for each run
Molecular vs. spore analyses and its influence on the results for Acaulospora
The genus Acaulospora, which is frequent in the rain forest at high elevations (Haug et al. 2019), was completely absent in samples from the dry forest. This result is in accordance with results from an Afromontane dry forest in Ethiopia (Wubet et al. 2003, 2006, 2009). In the studies conducted by Wubet et al. (2003, 2006, 2009), AMF were extracted from mycorrhizal samples and identified using cloning and Sanger sequencing, revealing a dominance of Glomus sequences but no representatives of Acaulosporaceae. Furthermore, Rodríguez-Echeverría et al. (2017) studied AMF communities in the Gorongosa National Park (Africa, Mozambique) from soil samples during the dry season using 454 pyrosequencing and found Acaulosporaceae only in a negligible amount (0.08% of sequences). The area sampled by Rodríguez-Echeverría et al. (2017) included grasslands, savannas miombo and dry forests. Due to the high-throughput sequencing method used by Rodríguez-Echeverría et al. (2017), significantly more VTs were found (101 AMF VTs in forests) than in Ecuador, but there is great agreement on the dominance of members of the Glomeraceae family. In contrast, studies from dry forests using spore morphology to identify AMF frequently find Acaulosporaceae and often in high numbers (Aguilar-Fernandéz et al. 2009, Carillo-Saucedo et al. 2018 Guadarrama et al. 2014, Sousa et al. 2018, Marinho et al. 2019 and Teixeira-Rios et al. 2018).
The reason for the discrepancy of Acaulosporaceae in spore vs. molecular samples remains unknown. Primer sensitivity and molecular methodology seem to be an unlikely reason because the same methods have revealed several Acaulosporaceae in other habitats (e.g., Haug et al. 2019). Furthermore, Wubet et al. (2003, 2006, 2009) and Rodríguez-Echeverría et al. (2017) used a different primer combination and method, and did not find Acaulosporaceae in their samples.
There is evidence that colonization strategies and abundance of extraradical mycelium, spores, and intraradical mycelium differ among AMF groups and between seasons (Hart and Reader 2002, Varela-Cervero et al. 2015). The study of Hart and Reader (2002) compared rate and extent of colonization by measuring percentage root colonization, root fungal biomass, soil hyphal length, and soil fungal biomass of different AMF groups including Acaulosporaceae and Glomaceae (now classified as Glomeraceae). Hart and Reader (2002) concluded that “Glomaceae isolates had high root colonization but low soil colonization, Gigasporaceae isolates showed the opposite trend whereas Acaulosporaceae isolates had low root and soil colonization.” Furthermore, the authors found that Glomeraceae colonized roots faster than Acaulosporaceae and Gigasporaceae and thought this likely to be caused by spore vs. hyphal colonization (Hart and Reader 2002).
We therefore think that it is reasonable to assume that during the dry season, Acaulosporaceae might be readily found in the soil as spores but not in mycorrhizal samples.
Our sampling design included soil samples, so why is it that we still did not detect Acaulosporaceae from soil samples? Hempel et al. (2007) reported that molecular analyses from soil samples have a bias towards hyphal material and might underestimate spore DNA.
In the future, it might be worthwhile to include spore analysis from the soil, in addition to molecular analyses in order to investigate the true AMF potential within an ecosystem. Likewise, to understand which AMF species are actively associated with plants during the dry season, it is necessary to conduct molecular studies on mycorrhizal samples instead of the spores in the soil.
Combining spore analyses with molecular methods would also make the comparison of AMF communities among different dry forests more feasible as many studies from South America are solely based on spore morphology.
AMF community of Laipuna in a global context
Studies of AMF communities in tropical dry forests have only been conducted so far in Mexico (Aguilar-Fernandéz et al. 2009, Carillo-Saucedo et al. 2018, Guadarrama et al. 2014), Brazil (Sousa et al. 2018, Marinho et al. 2019 and Teixeira-Rios et al. 2018), and Africa (Wubet et al. 2003, 2006, 2009). Albeit the difficulties in comparing spore results vs. molecular results (see paragraph above), all studies conclude a high dominance of Glomeraceae with few abundant species: in Brazil it is Glomus macrocarpum (Pagano et al. 2013, Sousa et al. 2018, Marinho et al. 2019); in Mexico it is Glomus aff. fasciculatum (Aguilar-Fernandéz et al. 2009, Carillo-Saucedo et al. 2018), Glomus sinuosum (Aguilar-Fernandéz et al. 2009), and Funneliformis geosporum (Carillo-Saucedo et al. 2018, Guadarrama et al. 2014). We confirm Glomus sinuosum (OTU R31, VT69) and Glomus aff. fasciculatum (OTU R1, VT113) to occur in Ecuador as well, but only Glomus sinuosum was dominant occurring in 39% of the samples. Of the 15 OTUs in the Ecuadorian dry forest that could be assigned to VTs, eleven were also found in the African dry forest of the Gorongosa National Park and eight in an Ethiopian dry forest (Wubet et al. 2009) (Fig. S3). All of the four frequent VTs of the Ecuadorian dry forest also occur in the Ethiopian dry forest (Wubet et al. 2009: Glomus NF02 = VT248, Glomus NF04 = VT69, Glomus NF10 = VT155, Glomus NF12 = VT166). An overlap of AMF communities between South America and Africa was also reported by Stürmer et al. (2018) who found many shared species between the South American and African continents (52%) and between the Neotropical and Afrotropic realms (53%). These concordances on two different continents underline the global distribution of AMF. Nevertheless, it should not be neglected that many OTUs/virtual taxa from dry forests of South Ecuador and Africa revealed to be new to science.
This makes the AMF communities of dry forests both in Ecuador and Africa unique and worth preserving.
Data availability
The sequences generated in this study are available in the NCBI Gen Bank (https://www.ncbi.nlm.nih.gov) under the accession numbers given in the text.
Code availability
Not applicable.
Change history
05 August 2021
A Correction to this paper has been published: https://doi.org/10.1007/s11557-021-01721-9
References
Aguilar-Fernandéz M, Jaramillo VJ, Varela-Fregoso L, Gavito ME (2009) Short-term consequences of slash-and-burn practices on the arbuscular mycorrhizal fungi of a tropical dry forest. Mycorrhiza 19:179–186
Aguirre ZM, Kvist LP, Sánchez OT (2006) Bosques seco en Ecuador y su diversidad. In: Moraes M, Øllgaard LP, Kvist LP, Borchsenius F, Balsev H (eds) Botánica Económica de los Andes Centrales. Universidad Mayor de San Andrés, La Paz, pp 162–187
Beck E, Makeschin F, Haubrich F, Richter M, Bendix J, Valerezo C (2008) The ecosystem (Reserva Biológica San Francisco). In: Beck E, Bendix J, Kottke I, Makeschin F, Mosandl R (eds) Gradients in a tropical mountain ecosystem of Ecuador. Ecological Studies, vol 198. Springer Verlag, Berlin, pp 137–148
Carillo-Saucedo SM, Gavito ME, Siddique I (2018) Arbuscular mycorrhizal fungal spore communities of a tropical dry forest ecosystem show resilience to land-use change. Fungal Ecol 32:29–39
da Silva IR, de Mello CMA, Ferreira Neto RA, da Silva DKA, de Melo AD, Oehl F, Maia LC (2014) Diversity of arbuscular mycorrhizal fungi along an environmental gradient in the Brazilian semiarid. Appl Soil Ecol 84:166–175
Davison J et al (2015) Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349:970–973
Davison J et al (2021) Temperature and pH define the realized niche space of arbuscular mycorrhizal fungi. New Phytol, in press. https://doi.org/10.1111/nph.17240
DRYFLOR, Banda-R K, Delgado-Salinas A, Dexter KG, Linares-Palomino R, Oliveira-Filho A, Prado D et al (2016) Plant diversity patterns in neotropical dry forests and their conservation implications. Science 353:1383–1387
Gai JP, Christie P, Feng G, Li XL (2006) Twenty years of research on community composition and species distribution of arbuscular mycorrhizal fungi in China: a review. Mycorrhiza 16:229–239
Göker M, Grimm GW, Auch AF, Aurahs R, Kucera M (2010) A clustering optimization strategy for molecular taxonomy applied to planktonic foraminifera SSU rDNA. Evol Bioinformatics Online 6:97–112
Guadarrama P, Castillo S, Ramos-Zapata JA, Hernández-Cuevas LV, Camargo-Ricalde SL (2014) Arbuscular mycorrhizal communities in changing environments: the effects of seasonality and anthropogenic disturbance in a seasonal dry forest. Pedobiologia 57:87–95
Hart MM, Reader RJ (2002) Taxonomic basis for variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytologist 153:335–344
Haug I, Setaro S, Suárez JP (2013) Reforestation sites show similar and nested AMF communities to an adjacent pristine forest in a tropical mountain area of South Ecuador. PLoS ONE 8:e63524
Haug I, Setaro S, Suárez JP (2019) Species composition of arbuscular mycorrhizal communities changes with elevation in the Andes of South Ecuador. PLoS ONE 14:e0221091
Hempel S, Renker C, Buscot F (2007) Differences in the species composition of arbuscular mycorrhizal fungal communities in spore, root and soil communities in a grassland ecosystem. Environ Microbiol 9:1930–1938
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Kottke I, Haug I, Setaro S, Suárez JP, Weiß M, Preußing M, Nebel M, Oberwinkler F (2008) Guilds of mycorrhizal fungi and their relation to trees, ericads, orchids and liverworts in a neotropical mountain rain forest. Basic Appl Ecol 9:13–23
Lee J, Lee S, Young JPW (2008) Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol Ecol 65:339–349
Lugo MA, Pagano MC (2019) Overview of the mycorrhizal fungi in South America. In: Pagano MC, Lugo MA (eds) Mycorrhizal Fungi in South America. Fungal Biology, Springer Nature, Switzerland, pp 1–27
Maitra P, Zheng Y, Chen L, Wang Y-L, Ji N-N, Lü P-P, Gan H-Y, Sun X-C, Zhou X-H, Guo L-D (2019) Effect of drought and season on arbuscular mycorrhizal fungi in a subtropical secondary forest. Fungal Ecol 41:107–115
Marinho F, da Silva IR, Oehl F, Maia LC (2018) Checklist of arbuscular mycorrhizal fungi in tropical forests. Sydowia 70:107–127. https://doi.org/10.12905/0380.sydowia70-2018-0107
Marinho F, Oehl F, Ramalho da Silva I, Coyne D, da Nóbrega Veras JS, Maia LC (2019) High diversity of arbuscular mycorrhizal fungi in natural and anthropized sites of a Brazilian tropical dry forest (Caatinga). Fungal Ecology 40:82–91
Miles L, Newton AC, DeFries RS, Ravilious C, May I, Blyth S, Kapos V, Gordon JE (2006) A global overview of the conservation status of tropical dry forests. J Biogeogr 33:491–505
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D et al (2018) Vegan: Community Ecology Package. Version 2:5–3
Öpik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM, Reier Ü, Zobel M (2010) The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol 188:223–241
Öpik M, Zobel M, Cantero JJ, Davison J, Facelli JM, Hiiesalu I et al (2013) Global sampling of plant roots expands the described molecular diversity of arbuscular mycorrhizal fungi. Mycorrhiza 23:411–430
Pagano MC, da Silva DK, da Silva GA, Maia LC (2019) Tropical dry forest compared to rainforest and associated ecosystems in Brazil. In: Pagano MC, Lugo MA (eds) Mycorrhizal Fungi in South America. Fungal Biology, Springer Nature, Switzerland, pp 177–192
Pagano MC, Zandavalli RB, Aráujo FS (2013) Biodiversity of arbuscular mycorrhizas in three vegetational types from the semiarid of Ceará State, Brazil. Appl Soil Ecol 67:37–46
Pucha-Cofrep D, Peters T, Bräuning A (2015) Wet season precipitation during the past century reconstructed from tree-rings of a tropical dry forest on Southern Ecuador. Glob Planet Chang 133:65–78
Rodríguez-Echeverría S, Teixeira H, Correia M, Timótes S, Heleno R, Öpik M, Moora M (2017) Arbuscular mycorrhizal fungi communities from tropical Africa reveal strong ecological structure. New Phytol 213:380–390
Sousa NMF, Veresoglou SD, Oehl F, Rillig MC, Maia LC (2018) Predictors of arbuscular mycorrhizal fungal communities in the Brazilian tropical dry forest. Microb Ecol 75:447–458
Stürmer SL, Bever JD, Morton JB (2018) Biogeography of arbuscular mycorrhizal fungi (Glomeromycota): a phylogenetic perspective on species distribution patterns. Mycorrhiza 28:587–603
Teixeira-Rios T, da Silva DKA, Goto BT, Yano-Melo AM (2018) Seasonal differences in arbuscular mycorrhizal fungal communities in two woody species dominating semiarid caatinga forests. Folia Geobot 53:191–200
Van Geel M, Busschaert P, Honnay O, Lievens B (2014) Evaluation of six primer pairs targeting the nuclear rRNA operon for characterization of arbuscular mycorrhizal fungal (AMF) communities using 454 pyrosequencing. J Microbiol Methods 106:93–100
Varela-Cervero S, Vasar M, Davison J, Barea JM, Öpik M, Azcón-Aguilar C (2015) The composition of arbuscular mycorrhizal fungal communities differs among the roots, spores and extraradical mycelia associated with five Mediterranean plant species. Environ Biol 17:2882–2895
White TJ, Bruns T, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: A guide to methods and application. Academic Press, New York, pp 315–322
Wubet T, Kottke I, Teketay D, Oberwinkler F (2003) Mycorrhizal status of indigenous trees in dry Afromontane forests of Ethiopia. For Ecol Manag 179:387–399
Wubet T, Kottke I, Teketay D, Oberwinkler F (2009) Arbuscular mycorrhizal community structures differ between co-occurring tree species of dry Afromontane tropical forest, and their seedlings exhibit potential to trap isolates suited for reforestation. Mycol Prog 8:317–328
Wubet T, Weiß M, Kottke I, Teketay D, Oberwinkler F (2006) Phylogenetic analysis of nearly small subunit rDNA sequences suggests that the endangered African Pencil Cedar, Juniperus procera, is associated with distinct members of Glomeraceae. Mycological Research 110:1059–1069
Acknowledgements
We thank Jutta Bloschies for field and laboratory work. We appreciate the Naturaleza y Cultura Internacional (Loja, San Diego) for support, our Ecuadorian partner Universidad Técnica Particular de Loja (UTPL) for outstanding cooperation, the Ecuadorian Ministerio del Ambiente for granting research permits, and the staff of the Estación Científica San Francisco—especially Felix Matt—for their great assistance.
Funding
Open Access funding enabled and organized by Projekt DEAL. The study was funded by the Deutsche Forschungsgemeinschaft within the Research Units 402 and 816 and PAK825.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Ingeborg Haug and Sabrina Setaro. The first draft of the manuscript was written by Ingeborg Haug, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Marco Thines
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised. Figure 3 to 5 captions are now corrected.
Supplementary Information
Table S1
Accession numbers, Notations, Isolation Sources and OTU Numbers of the Sequences (XLSX 57 kb)
Fig. S1
Map of the Study Region in South Ecuador Location of the study sites: Laipuna dry forest (600 – 1200 masl), Bombuscaro evergreen premontane forest (1000 masl), RBSF evergreen lower montane forest (2000 masl), Cajanuma upper montane forest (3000 masl), Cajas grass páramo/Polylepis forest (4000 masl). http://vhrz669.hrz.uni-marburg.de/tmf_respect/data_pre.do?citid=1745 (PNG 7643 kb)
Fig. S2
Rarefaction Analysis for Sites in the Dry and Rain Forest (PNG 80 kb)
Fig. S3
Shared and Unique OTUs of the Laipuna Dry Forest with Respect to tropical Africa (Rodríguez-Echeverría et al. 2017) and the Elevation Belts of the Rain Forest The horizontal bars connecting the same sites indicate the amount of unique OTUs for the site. The thickness of connector lines between different sites represents the percentage of OTUs shared. Laipuna had a high number of shared OTUs with the tropical Africa. There are low numbers of shared OTUs between tropical Africa and the Ecuadorian rain forest. (PNG 242 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Haug, I., Setaro, S. & Suárez, J.P. Global AM fungi are dominating mycorrhizal communities in a tropical premontane dry forest in Laipuna, South Ecuador. Mycol Progress 20, 837–845 (2021). https://doi.org/10.1007/s11557-021-01699-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-021-01699-4