Abstract
Purpose
Neuroendocrine tumors (NETs) are a rare form of cancer that can occur anywhere in the body and commonly metastasizes. The large variance in location and aggressiveness of the tumors makes it a difficult cancer to treat. Assessments of the whole-body tumor burden in a patient image allow for better tracking of disease progression and inform better treatment decisions. Currently, radiologists rely on qualitative assessments of this metric since manual segmentation is unfeasible within a typical busy clinical workflow.
Methods
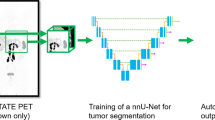
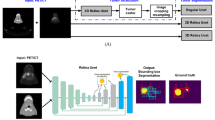
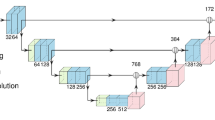
We address these challenges by extending the application of the nnU-net pipeline to produce automatic NET segmentation models. We utilize the ideal imaging type of 68Ga-DOTATATE PET/CT to produce segmentation masks from which to calculate total tumor burden metrics. We provide a human-level baseline for the task and perform ablation experiments of model inputs, architectures, and loss functions.
Results
Our dataset is comprised of 915 PET/CT scans and is divided into a held-out test set (87 cases) and 5 training subsets to perform cross-validation. The proposed models achieve test Dice scores of 0.644, on par with our inter-annotator Dice score on a subset 6 patients of 0.682. If we apply our modified Dice score to the predictions, the test performance reaches a score of 0.80.
Conclusion
In this paper, we demonstrate the ability to automatically generate accurate NET segmentation masks given PET images through supervised learning. We publish the model for extended use and to support the treatment planning of this rare cancer.



Similar content being viewed by others
References
Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, Shih T, Yao JC (2017) Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncol 3(10):1335–1342
Fallahi B, Manafi-Farid R, Eftekhari M, Fard-Esfahani A, Emami-Aderkani A, Geramifar P, Akhlaghi M, Taheri APH, Beiki D (2019) Diagnostic efficiency of 68Ga-DOTATATE PET/CT as compared to 99mTc-octreotide SPECT/CT and conventional morphologic modalities in neuroendocrine tumors. Asia Ocean J Nucl Med Biol. 7:129–140
Dromain C, Pavel ME, Ruszniewski P, Langley A, Massien C, Baudin E, Caplin ME (2019) Tumor growth rate as a metric of progression, response, and prognosis in pancreatic and intestinal neuroendocrine tumors. BMC Cancer 19(66)
Egger J, Kapur T, Fedorov A, Pieper S, Miller JV, Veeraraghavan H, Freisleben B, Golby AJ, Nimsky C, Kikinis R (2013) GBM volumetry using the 3D slicer medical image computing platform. Sci Rep 3:1364 https://doi.org/10.1038/srep01364
Antonelli M, Reinkeb A, Bakase S, Farahanif K, Kopp-Schneiderg A, Landman BA, Litjens G, Menze B, Ronneberger O, Summers RM, Van Ginneken B, Bilello M, Bilic P, Christ PF, Do RKG, Gollub MJ, Heckers SH, Huisman H, Jarnagin WR, McHugo MK, Napel S, Pernicka JSG, Rohde K, Tonbo-Gomex C, Vorontsov E, Huisman H, Meakin JA, Ourselin S, Wiesenfarth M, Arbelaex P, Bae B, Chen S, Daza L, Feng J, He B, Isensee F, Ji Y, Jia F, Kim N, Kim I, Merhof D, Pai A, Park B, Perslev M, Rezaiifar R, Rippel O, Sarasua I, Shen W, Son J, Wachinger C, Wang L,Wang Y, Xia Y, Xu D, Xu Z, Zheng Y, Simpson AL, Maier-Hein L, Jorge Cardoso M (2021) The medical segmentation decathlon. Nat Digit Med Nat Commun 13:4128. https://doi.org/10.1038/s41467-022-30695-9
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. Med Image Comput Comput Assist Interv MICCAI
Yousefirizi F, Jha AK, Brosch-Lenz J, Saboury B, Rahmin A (2021) Towards high-throughput artificial intelligence-base segmentation in oncological pet imaging. PET Clin 16:577–596
Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18:203–211
Warfield SK, Zou KH, Wells WM (2004) Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 23(7):903–921
Huang H, Lin L, Tong R, Hu H, Zhang Q, Iwamoto Y, Han X, Chen YW, Wu J (2015) Unet 3+: A full-scale connected Unet for medical image segmentation. In: IEEE international conference on acoustics, speech and signal processing (ICASSP), pp 1055–1059
Kamnitsas K, Chen L, Ledig C, Rueckert D, Glocker B (2015) Multi-scale 3D CNNs for segmentation of brain lesions in multi-modal MRI. Med Image Comput Comput Assist Intervent MICCAI
Karimi D, Dou H, Warfield SK, Gholipour S (2020) Deep learning with noisy labels: exploring techniques and remedies in medical image analysis. Med Image Anal 65
Funding
This project was supported by the National Institutes of Health and National Cancer Institute (P30 CA008748).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest. Pierre Elnajjar currently employed by Regeneron, Inc.
Ethical approval:
This retrospective study was approved by the local institutional review board, and the need for written informed consent was waived. All data storage and handling were performed in compliance with Health Insurance Portability and Accountability Act regulations.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Link to information about the trained model: https://github.com/AliceSantilli/nnUNet.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Santilli, A., Panyam, P., Autz, A. et al. Automated full body tumor segmentation in DOTATATE PET/CT for neuroendocrine cancer patients. Int J CARS 18, 2083–2090 (2023). https://doi.org/10.1007/s11548-023-02968-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-023-02968-1