Abstract
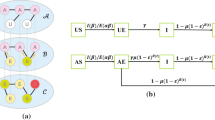
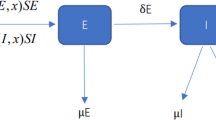
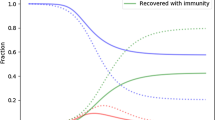
Due to the complex interactions between multiple infectious diseases, the spreading of diseases in human bodies can vary when people are exposed to multiple sources of infection at the same time. Typically, there is heterogeneity in individuals' responses to diseases, and the transmission routes of different diseases also vary. Therefore, this paper proposes an SIS disease spreading model with individual heterogeneity and transmission route heterogeneity under the simultaneous action of two competitive infectious diseases. We derive the theoretical epidemic spreading threshold using quenched mean-field theory and perform numerical analysis under the Markovian method. Numerical results confirm the reliability of the theoretical threshold and show the inhibitory effect of the proportion of fully competitive individuals on epidemic spreading. The results also show that the diversity of disease transmission routes promotes disease spreading, and this effect gradually weakens when the epidemic spreading rate is high enough. Finally, we find a negative correlation between the theoretical spreading threshold and the average degree of the network. We demonstrate the practical application of the model by comparing simulation outputs to temporal trends of two competitive infectious diseases, COVID-19 and seasonal influenza in China.










Similar content being viewed by others
References
Albert R, Barabási AL (2002) Statistical mechanics of complex networks. Rev Mod Phys 74(1):47
Anderson RM, May RM (1991) Infectious diseases of humans: dynamics and control. Oxford University Press
Barabási AL, Albert R (1999) Emergence of scaling in random networks. Science 286(5439):509–512
Biggerstaff M, Cauchemez S, Reed C, Gambhir M, Finelli L (2014) Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infect Dis 14(1):1–20
Boguná M, Castellano C, Pastor-Satorras R (2013) Nature of the epidemic threshold for the susceptible-infected-susceptible dynamics in networks. Phys Rev Lett 111(6):068701
Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M (2007) Transmission of influenza A in human beings. Lancet Infect Dis 7(4):257–265
Cai CR, Wu ZX, Chen MZ, Holme P, Guan JY (2016) Solving the dynamic correlation problem of the susceptible-infected-susceptible model on networks. Phys Rev Lett 116(25):258301
Chen J, Gu C, Ruan Z, Tang M (2023) Competition of SARS-CoV-2 variants on the pandemic transmission dynamics. Chaos Solitons Fractals 169:113193
Chinese Center for Disease Control and Prevention (China CDC) https://www.chinacdc.cn/
Colizza V, Vespignani A (2007) Invasion threshold in heterogeneous metapopulation networks. Phys Rev Lett 99(14):148701
Colizza V, Pastor-Satorras R, Vespignani A (2007) Reaction–diffusion processes and metapopulation models in heterogeneous networks. Nat Phys 3(4):276–282
Dhanasekaran V, Sullivan S, Edwards KM, Xie R, Khvorov A, Valkenburg SA, Barr IG (2022) Human seasonal influenza under COVID-19 and the potential consequences of influenza lineage elimination. Nat Commun 13(1):1721
Eames KT, Keeling MJ (2002) Modeling dynamic and network heterogeneities in the spread of sexually transmitted diseases. Proc Natl Acad Sci 99(20):13330–13335
Erdős P, Rényi A (1960) On the evolution of random graphs. Publ Math Inst Hung Acad Sci 5(1):17–60
Fang F, Ma J, Li Y (2023) The coevolution of the spread of a disease and competing opinions in multiplex networks. Chaos Solitons Fractals 170:113376
Gao B, Deng Z, Zhao D (2016) Competing spreading processes and immunization in multiplex networks. Chaos Solitons Fractals 93:175–181
Gormley M, Marawska L, Milton D (2020) It is time to address airborne transmission of coronavirus disease 2019 (COVID-19). Clin Infect Dis 71(9):2311–2313
Granell C, Gómez S, Arenas A (2013) Dynamical interplay between awareness and epidemic spreading in multiplex networks. Phys Rev Lett 111(12):128701
He R, Luo X, Asamoah JKK, Zhang Y, Li Y, Jin Z, Sun GQ (2023) A hierarchical intervention scheme based on epidemic severity in a community network. J Math Biol 87(2):29
Karrer B, Newman ME (2011) Competing epidemics on complex networks. Phys Rev E 84(3):036106
Kim H, Ha M, Jeong H (2015) Scaling properties in time-varying networks with memory. Eur Phys J B 88:1–8
Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J (2020) The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. https://doi.org/10.1093/jtm/taaa021
Luo XF, Feng S, Yang J, Peng XL, Cao X, Zhang J, Jin Z (2021) Nonpharmaceutical interventions contribute to the control of COVID-19 in China based on a pairwise model. Infect Dis Modell 6:643–663
Meyers L (2007) Contact network epidemiology: Bond percolation applied to infectious disease prediction and control. Bull Am Math Soc 44(1):63–86
Newman ME (2002) Spread of epidemic disease on networks. Phys Rev E 66(1):016128
Nie Y, Li W, Pan L, Lin T, Wang W (2022) Markovian approach to tackle competing pathogens in simplicial complex. Appl Math Comput 417:126773
Parshani R, Carmi S, Havlin S (2010) Epidemic threshold for the susceptible-infectious-susceptible model on random networks. Phys Rev Lett 104(25):258701
Pastor-Satorras R, Vespignani A (2001a) Epidemic dynamics and endemic states in complex networks. Phys Rev E 63(6):066117
Pastor-Satorras R, Vespignani A (2001b) Epidemic spreading in scale-free networks. Phys Rev Lett 86(14):3200
Pastor-Satorras R, Castellano C, Van Mieghem P, Vespignani A (2015) Epidemic processes in complex networks. Rev Mod Phys 87(3):925
Perra N, Gonçalves B, Pastor-Satorras R, Vespignani A (2012) Activity driven modeling of time varying networks. Sci Rep 2(1):469
Rand DA (1999) Correlation equations and pair approximations for spatial ecologies. In: McGlade J (ed) Advanced ecological theory: principles and applications. Wiley, pp 100–142. https://doi.org/10.1002/9781444311501.ch4
Rodriguez-Barraquer I, Costa F, Nascimento EJ, Nery N, Castanha PM, Sacramento GA, Ko AI (2019) Impact of preexisting dengue immunity on Zika virus emergence in a dengue endemic region. Science 363(6427):607–610
Sahneh FD, Scoglio C (2014) Competitive epidemic spreading over arbitrary multilayer networks. Phys Rev E 89(6):062817
Sanche S, Lin YT, Xu C, Romero-Severson E, Hengartner N, Ke R (2020) High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26(7):1470
Sanz J, Xia CY, Meloni S, Moreno Y (2014) Dynamics of interacting diseases. Phys Rev X 4(4):041005
Saumell-Mendiola A, Serrano MÁ, Boguná M (2012) Epidemic spreading on interconnected networks. Phys Rev E 86(2):026106
Soriano-Paños D, Ghanbarnejad F, Meloni S, Gómez-Gardeñes J (2019) Markovian approach to tackle the interaction of simultaneous diseases. Phys Rev E 100(6):062308
Sun Q, Wang Z, Zhao D, Xia C, Perc M (2022) Diffusion of resources and their impact on epidemic spreading in multilayer networks with simplicial complexes. Chaos Solitons Fractals 164:112734
Wang J, Du G (2020) COVID-19 may transmit through aerosol. Ir J Med Sci 1971- 189:1143–1144
Wang W, Tang M, Stanley HE, Braunstein LA (2017) Unification of theoretical approaches for epidemic spreading on complex networks. Rep Prog Phys 80(3):036603
Watts DJ, Strogatz SH (1998) Collective dynamics of ‘small-world’ networks. Nature 393(6684):440–442
Yang JX (2019) Epidemic spreading on multilayer homogeneous evolving networks. Chaos Interdiscip J Nonlinear Sci 29(10):103146
Zhao Y, Zheng M, Liu Z (2014) A unified framework of mutual influence between two pathogens in multiplex networks. Chaos Interdiscip J Nonlinear Sci 24(4):043129
Zipfel CM, Colizza V, Bansal S (2021) The missing season: the impacts of the COVID-19 pandemic on influenza. Vaccine 39(28):3645–3648
Acknowledgements
The authors would like to thank the anonymous referees for their valuable comments and suggestions, which have effectively improved the quality of this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, X., Xu, Y. Markovian Approach for Exploring Competitive Diseases with Heterogeneity-Evidence from COVID-19 and Influenza in China. Bull Math Biol 86, 71 (2024). https://doi.org/10.1007/s11538-024-01300-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-024-01300-5