Abstract
To characterize Coronavirus Disease 2019 (COVID-19) transmission dynamics in each of the metropolitan statistical areas (MSAs) surrounding Dallas, Houston, New York City, and Phoenix in 2020 and 2021, we extended a previously reported compartmental model accounting for effects of multiple distinct periods of non-pharmaceutical interventions by adding consideration of vaccination and Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) variants Alpha (lineage B.1.1.7) and Delta (lineage B.1.617.2). For each MSA, we found region-specific parameterizations of the model using daily reports of new COVID-19 cases available from January 21, 2020 to October 31, 2021. In the process, we obtained estimates of the relative infectiousness of Alpha and Delta as well as their takeoff times in each MSA (the times at which sustained transmission began). The estimated infectiousness of Alpha ranged from 1.1x to 1.4x that of viral strains circulating in 2020 and early 2021. The estimated relative infectiousness of Delta was higher in all cases, ranging from 1.6x to 2.1x. The estimated Alpha takeoff times ranged from February 1 to February 28, 2021. The estimated Delta takeoff times ranged from June 2 to June 26, 2021. Estimated takeoff times are consistent with genomic surveillance data.






Similar content being viewed by others
Notes
Department of Health and Human Services. COVID-19 vaccine milestones [cited 2022 Dec 21]. https://www.hhs.gov/coronavirus/covid-19-vaccines/distribution/index.html
Centers for Disease Control and Prevention. COVID data tracker. 2021 [cited 2021 Oct 6] https://covid.cdc.gov/covid-data-tracker/#datatracker-home
Coronavirus Resource Center, Johns Hopkins University. Impact of opening and closing decisions by state: a look at how social-distancing measures may have influenced trends in COVID-19 cases and deaths. 2021 [cited 2021 Oct 6]. https://coronavirus.jhu.edu/data/state-timeline
Centers for Disease Control and Prevention. Covid data tracker, variant proportions. 2021 [cited 2021 Oct 6]. https://covid.cdc.gov/covid-data-tracker/#variant-proportions
Public Health England. SARS-CoV-2 variants of concern and variants under investigation in England. Technical briefing 15, June 11, 2021. [cited 2021 Aug 24]. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/993879/Variants_of_Concern_VOC_Technical_Briefing_15.pdf
The New York Times. Coronavirus (COVID-19) data in the United States. 2021 [cited 2021 Aug 24]. https://github.com/nytimes/covid-19-data
Covid Act Now. US COVID risk and vaccine tracker. 2021 [cited 2021 Sep 29]. https://covidactnow.org/
Democrat and Chronicle. COVID-19 vaccine tracker. 2023 [cited 2023 Nov 11]. https://data.democratandchronicle.com/covid-19-vaccine-tracker
GitHub repository of supplemental materials for this study. 2023 [cited 2023 Dec 13]. https://github.com/lanl/PyBNF/tree/master/examples/Vax_and_Variants
Centers for Disease Control and Prevention. SARS-CoV-2 variant proportions. 2023 [cited 2023 Mar 8]. https://data.cdc.gov/Laboratory-Surveillance/SARS-CoV-2-Variant-Proportions/jr58-6ysp
References
Alexander TY, Hughes B, Wolfe MK, Leon T, Duong D, Rabe A et al (2022) Estimating relative abundance of 2 SARS-CoV-2 variants through wastewater surveillance at 2 large metropolitan sites. United States Emerg Infect Dis 28:940. https://doi.org/10.3201/eid2805.212488
Andre M, Lau LS, Pokharel MD, Ramelow J, Owens F, Souchak J et al (2023) From Alpha to Omicron: How different variants of concern of the SARS-Coronavirus-2 impacted the world. Biology 12:1267. https://doi.org/10.3390/biology12091267
Andrieu C, Thoms J (2008) A tutorial on adaptive MCMC. Stat Comput 18:343–373. https://doi.org/10.1007/s11222-008-9110-y
Arav Y, Fattal E, Klausner Z (2022) Increased transmissibility of emerging SARS-CoV-2 variants is driven either by viral load or probability of infection rather than environmental stability. Mathematics 10(19):3422. https://doi.org/10.3390/math10193422
Barnard RC, Davies NG, Jit M, Edmunds WJ (2022) Modeling the medium-term dynamics of SARS-CoV-2 transmission in England in the Omicron era. Nat Commun 13:4879. https://doi.org/10.1038/s41467-022-32404-y
Bo Y, Guo C, Lin C, Zeng Y, Li HB et al (2021) Effectiveness of non-pharmaceutical interventions on COVID-19 transmission in 190 countries from 23 January to 13 April 2020. Int J Inf Dis 102:247–253. https://doi.org/10.1016/j.ijid.2020.10.066
Bolze A, Luo S, White S, Cirulli ET, Wyman D, Dei Rossi A et al (2022) SARS-CoV-2 variant Delta rapidly displaced variant Alpha in the United States and led to higher viral loads. Cell Rep Med. https://doi.org/10.1016/j.xcrm.2022.100564
Cai J, Deng X, Yang J, Sun K, Liu H, Chen Z et al (2022) Modeling transmission of SARS-CoV-2 Omicron in China. Nat Med 28:1468–1475. https://doi.org/10.1038/s41591-022-01855-7
Campbell F, Archer B, Laurenson-Schafer H, Jinnai Y, Konings F, Batra N et al (2021) Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill 26:2100509. https://doi.org/10.2807/1560-7917.es.2021.26.24.2100509
Cascella M, Rajnik M, Aleem A, Dulebohn SC, Di Napoli R (2022) Features, evaluation, and treatment of coronavirus (COVID-19). July 30, 2021 update. Treasure Island (FL): StatPearls Publishing. https://www.ncbi.nlm.nih.gov/books/NBK554776/
Chand M et al (2024) Investigation of novel SARS-COV-2 variant: variant of Concern 202012/01. Technical Briefing 1. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959438/Technical_Briefing_VOC_SH_NJL2_SH2.pdf (Public Health England, 2020)
Courtemanche C, Garuccio J, Le A, Pinkston J, Yelowitz A (2020) Strong social-distancing measures in the United States reduced the COVID-19 growth rate. Health Aff (Millwood) 39:1237–1246. https://doi.org/10.1377/hlthaff.2020.00608
Daniel W, Nivet M, Warner J, Podolsky DK (2021) Early evidence of the effect of SARS-CoV-2 vaccine at one medical center. N Engl J Med 384:1962–1963. https://doi.org/10.1056/NEJMc2102153
Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD et al (2021a) Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372:eabg3055. https://doi.org/10.1126/science.abg3055
Davies NG, Jarvis CI, Edmunds WJ, Jewell NP, Diaz-Ordaz K, Keogh RH (2021b) Increased mortality in community-tested cases of SARS-CoV-2 lineage B. 1.1. 7. Nature 593:270–274. https://doi.org/10.1038/s41586-021-03426-1
Donnelly MA, Chuey MR, Soto R, Schwartz NG, Chu VT, Konkle SL et al (2022) Household transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Alpha variant–United States, 2021. Clin Infect Dis 75:e122–e132. https://doi.org/10.1093/cid/ciac125
Earnest R, Uddin R, Matluk N, Renzette N, Turbett SE, Siddle KJ et al (2022) Comparative transmissibility of SARS-CoV-2 variants delta and alpha in New England, USA. Cell Rep Med. https://doi.org/10.1016/j.xcrm.2022.100583
Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A et al (2021) Emergence of SARS-CoV-2 B.1.1.7 lineage—United States, December 29, 2020–January 12, 2021. MMWR Morb Mortal Wkly Rep 70(3):95–99. https://doi.org/10.15585/mmwr.mm7003e2
Herlihy R, Bamberg W, Burakoff A, Alden N, Severson R, Bush E et al (2021) Rapid increase in circulation of the SARS-CoV-2 B.1.617.2 (Delta) variant—Mesa County, Colorado, April–June 2021. MMWR Morb Mortal Wkly Rep 70:1084–1087. https://doi.org/10.15585/mmwr.mm7032e2
Hindmarsh AC (1983) ODEPACK, a systematized collection of ODE solvers. In: Stepleman RS (ed) Scientific computing: applications of mathematics and computing to the physical sciences. North-Holland Publishing Company, Amsterdam, pp 55–64
Hsiang S, Allen D, Annan-Phan S, Bell K, Bolliger I, Chong T et al (2020) The effect of large-scale anti-contagion policies on the COVID-19 pandemic. Nature 584:262–267. https://doi.org/10.1038/s41586-020-2404-8
Ito K, Piantham C, Nishiura H (2021) Predicted dominance of variant Delta of SARS-CoV-2 before Tokyo Olympic Games, Japan, July 2021. Euro Surveill 26:2100570. https://doi.org/10.2807/1560-7917.ES.2021.26.27.2100570
Korodi M, Rákosi K, Jenei Z, Hudák G, Horváth I, Kákes M et al (2022) Longitudinal determination of mRNA-vaccination induced strongly binding SARS-CoV-2 IgG antibodies in a cohort of healthcare workers with and without prior exposure to the novel coronavirus. Vaccine 40(37):5445–5451. https://doi.org/10.1016/j.vaccine.2022.07.040
Lin YT, Neumann J, Miller EF, Posner RG, Mallela A, Safta C, Ray J, Thakur G, Chinthavali S, Hlavacek WS (2021) Daily forecasting of regional epidemics of Coronavirus disease with Bayesian uncertainty quantification, United States. Emerg Infect Dis 27:767–778. https://doi.org/10.3201/eid2703.203364
Liossi S, Tsiambas E, Maipas S, Papageorgiou E, Lazaris A, Kavantzas N (2023) Mathematical modeling for Delta and Omicron variant of SARS-CoV-2 transmission dynamics in Greece. Infect Dis Model 8:794–805. https://doi.org/10.1016/j.idm.2023.07.002
Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S et al (2021) Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) variant. N Engl J Med 385:585–594. https://doi.org/10.1056/NEJMoa2108891
Mallela A, Neumann J, Miller EF, Chen Y, Posner RG, Lin YT, Hlavacek WS (2022) Bayesian inference of state-level COVID-19 basic reproduction numbers across the United States. Viruses 14:157. https://doi.org/10.3390/v14010157
Mallela A, Lin YT, Hlavacek WS (2023) Differential contagiousness of respiratory disease across the United States. Epidemics 45:100718. https://doi.org/10.1016/j.epidem.2023.100718
Matrajt L, Leung T (2020) Evaluating the effectiveness of social-distancing interventions to delay or flatten the epidemic curve of coronavirus disease. Emerg Infect Dis 26:1740–1748. https://doi.org/10.3201/eid2608.201093
Michaelsen TY, Bennedbæk M, Christiansen LE, Jørgensen MS, Møller CH, Sørensen EA et al (2022) Introduction and transmission of SARS-CoV-2 lineage B.1.1.7, Alpha variant, in Denmark. Genome Med. 14:47. https://doi.org/10.1186/s13073-022-01045-7
Neumann J, Lin YT, Mallela A, Miller EF, Colvin J, Duprat AT, Chen Y, Hlavacek WS, Posner RG (2022) Implementation of a practical Markov chain Monte Carlo sampling algorithm in PyBioNetFit. Bioinformatics 38:1770–1772. https://doi.org/10.1093/bioinformatics/btac004
Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M et al (2020) The Advisory Committee on Immunization Practices’ interim recommendation for use of Pfizer-BioNTech COVID-19 vaccine—United States, December 2020. MMWR Morb Mortal Wkly Rep 69:1922–1924. https://doi.org/10.15585/mmwr.mm6950e2
Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M et al (2021) The Advisory Committee on Immunization Practices’ interim recommendation for use of Moderna COVID-19 vaccine—United States, December 2020. MMWR Morb Mortal Wkly Rep 69:1653–1656. https://doi.org/10.15585/mmwr.mm695152e1
Puranik A, Lenehan PJ, Silvert E, Niesen MJM, Corchado-Garcia J, O’Horo JC et al (2024) Comparison of two highly-effective mRNA vaccines for COVID-19 during periods of Alpha and Delta variant prevalence. [cited 2021 Oct 8]. https://doi.org/10.1101/2021.08.06.21261707v3
Rovida F, Cassaniti I, Paolucci S, Percivalle E, Sarasini A, Piralla A et al (2021) SARS-CoV-2 vaccine breakthrough infections with the alpha variant are asymptomatic or mildly symptomatic among health care workers. Nat Commun 12:6032. https://doi.org/10.1038/s41467-021-26154-6
Sheikhi F, Yousefian N, Tehranipoor P, Kowsari Z (2022) Estimation of the basic reproduction number of Alpha and Delta variants of COVID-19 pandemic in Iran. PLoS ONE 17:e0265489. https://doi.org/10.1371/journal.pone.0265489
Sonabend R, Whittles LK, Imai N, Perez-Guzman PN, Knock ES, Rawson T et al (2021) Non-pharmaceutical interventions, vaccination, and the SARS-CoV-2 delta variant in England: a mathematical modelling study. Lancet 398:1825–1835. https://doi.org/10.1016/s0140-6736(21)02276-5
Stein C, Nassereldine H, Sorensen RJ, Amlag JO, Bisignano C, Byrne S et al (2023) Past SARS-CoV-2 infection protection against re-infection: a systematic review and meta-analysis. Lancet 401:833–842. https://doi.org/10.1016/S0140-6736(22)02465-5
Tabatabai M, Juarez PD, Matthews-Juarez P, Wilus DM, Ramesh A, Alcendor DJ et al (2023) An analysis of COVID-19 mortality during the dominancy of Alpha, Delta and Omicron in the USA. J Prim Care Community Health 14:21501319231170164. https://doi.org/10.1177/21501319231170164
Tang P, Hasan MR, Chemaitelly H, Yassine HM, Benslimane FM, Al Khatib HA et al (2021) BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 Delta variant in Qatar. Nat Med 27:2136–2143
Thompson MG, Burgess JL, Naleway AL, Tyner HL, Yoon SK, Meece J et al (2021) Interim estimates of vaccine effectiveness of BNT162b2 and mRNA-1273 COVID-19 vaccines in preventing SARS-CoV-2 infection among health care personnel, first responders, and other essential and frontline workers—eight U.S. locations, December 2020–March 2021. MMWR Morb Mortal Wkly Rep 70:495–500. https://doi.org/10.15585/mmwr.mm7013e3
Vats D, Knudson C (2021) Revisiting the Gelman-Rubin Diagnostic. Statist Sci 36:518–529. https://doi.org/10.1214/20-STS812
Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L et al (2021) Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593:266–269. https://doi.org/10.1038/s41586-021-03470-x
Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Schiabor Barrett KM et al (2021) Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell 184:2587–2594. https://doi.org/10.1016/j.cell.2021.03.052
Zárate S, Taboada B, Muñoz-Medina JE, Iša P, Sanchez-Flores A, Boukadida C et al (2022) The alpha variant (B. 1.1. 7) of SARS-CoV-2 failed to become dominant in Mexico. Microbiol Spectr 10:e02240-e2321. https://doi.org/10.1128/spectrum.02240-21
Acknowledgements
W.S.H., Y.T.L., and A.M. were supported by the LDRD program at Los Alamos National Laboratory (Project 20220268ER). Y.C., Z.H., E.F.M., J.N., K.E.N., and R.G.P. were supported by a grant from the National Institute of General Medical Sciences of the National Institutes of Health (Grant R01GM111510). A.M. was supported by the 2020 Mathematical Sciences Graduate Internship program, which is sponsored by the Division of Mathematical Sciences of the National Science Foundation, and the Center for Nonlinear Studies at Los Alamos National Laboratory. Computational resources used in this study included Northern Arizona University’s Monsoon computer cluster, which is funded by Arizona’s Technology and Research Initiative Fund, and the FARM computer cluster at the University of California, Davis. Y.C. thanks Song Chen (University of Wisconsin, La Crosse, Wisconsin, USA) for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
1.1 Imputation of Missing Daily Case Counts
By October 31, 2021, many regions in the US were not reporting new detected COVID-19 cases on a strictly daily basis. When one or more daily case counts were not available, we imputed daily case counts on the basis of a linear fit to the two nearest available cumulative case counts. This approach has the effect of evenly distributing case counts across the days for which daily reports are unavailable.
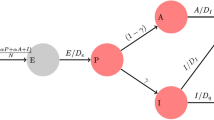
1.2 Equations of the Compartmental Model
The compartmental model, which is illustrated in Fig. 1 and Fig. 7 in Appendix, consists of the following 40 ordinary differential equations (ODEs):
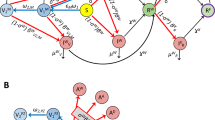
Expanded illustration of the new compartmental model. In the extended model, vaccination was considered by allowing susceptible and recovered persons to transition into a vaccinated compartment, either \({V}_{1}\) and \({R}_{V}\). Susceptible (recovered) persons who have completed vaccination move into the \({V}_{1}\) (\({R}_{V}\)) compartment. The susceptible persons who move into \({V}_{1}\) are drawn from \({S}_{M}\) (populated by susceptible persons who are mixing and unprotected by social-distancing) and from \({S}_{P}\) (populated by susceptible persons who are protected by social-distancing). After susceptible persons enter \({V}_{1}\), they can move through a series of additional compartments (\({V}_{2}\) through \({V}_{6}\)), which are included to capture the time needed for immunity to develop after completion of vaccination. We estimate that the time needed to acquire immunity after vaccination is approximately three weeks based on longitudinal studies of anti-spike protein IgG levels (Bo et al. 2021). Persons who exit the \({V}_{6}\) compartment without becoming infected enter one of the following compartments: \({S}_{V,1}\), \({S}_{V,2}\), \({S}_{V,3}\), or \({S}_{V,4}\). Persons in \({S}_{V,1}\) are taken to remain susceptible to productive infection by all SARS-CoV-2 strains of interest (Alpha, Delta, and ancestral strains). Persons in \({S}_{V,2}\) are taken to be susceptible to SARS-CoV-2 Alpha and Delta variants. Persons in \({S}_{V,3}\) are taken to be susceptible to Delta. Persons in \({S}_{V,4}\) are taken to be protected against all strains of interest. Infection of persons in \({S}_{V,3}\) is only allowed if Delta is present, i.e., at times \(t>{\theta }_{2}\). Infection of persons in \({S}_{V,2}\) is only allowed if Alpha or Delta is present, i.e., at times \(t>{\theta }_{1}\). Vaccinated persons in compartments \({V}_{1}\) through \({V}_{6}\) and compartment \({S}_{V,1}\) are allowed to become infected at any time, at which point they transition to compartment \({E}_{V}\), consisting of vaccinated persons who were exposed before development of vaccine-induced immunity. Persons in compartment \({S}_{V,2}\) are allowed to become infected if \(t\ge {\theta }_{1}\). Similarly, persons in compartment \({S}_{V,3}\) are allowed to become infected if \(t\ge {\theta }_{2}\). Possible outcomes for persons in \({E}_{V}\) are taken to be the same as those for unvaccinated exposed persons; however, the incubation period is taken to be distinct. Persons in \({E}_{V}\) can experience asymptomatic disease (upon entering \({A}_{V})\) or they can become symptomatic (upon entering \({I}_{V}\)). Persons in \({A}_{V}\) eventually recover, entering compartment \({R}_{V}\). Persons in \({I}_{V}\) can progress to severe disease (upon entering \({H}_{V}\)) or recover (upon entering \({R}_{V}\)). Persons in \({H}_{V}\) either recover (moving into \({R}_{V}\)) or die (moving into \(D\)). Persons who have recovered from infection, in the \({R}_{U}\) compartment, move directly into the \({R}_{V}\) compartment upon vaccination. Persons in the \({R}_{U}\) and \({R}_{V}\) compartments are taken to have full immunity. The vaccination rate at which susceptible and recovered persons move into vaccinated compartments is updated daily for consistency with the empirical overall rate of vaccination, which we extract daily from the COVID Act Now Data API (Courtemanche et al. 2020) and the Democrat and Chronicle newspaper (Hsiang et al. 2020). The relative values of the vaccination rate are set such that each person eligible for vaccination has the same probability of being vaccinated. All unvaccinated persons are taken to be eligible for vaccination except symptomatic persons (in compartments \({I}_{M}\) and \({I}_{P}\)), persons who are hospitalized or severely ill at home (in compartment \(H\)), quarantined persons (in the various compartments labeled with a Q subscript), and deceased persons (in compartment \(D\)). It should be noted that asymptomatic, non-quarantined persons (in compartments \(A{}_{M}\) and \({A}_{P}\)) and presymptomatic, non-quarantined persons (in the \(E\) compartments) are taken to be eligible (and to influence the vaccination rate constants) but, as a simplification, these persons are not explicitly tracked as vaccinated or unvaccinated because each of these persons will eventually enter either the \(D\) compartment or the \({R}_{U}\) compartment, at which point they will have immunity. In the model, the effects of SARS-CoV-2 variants are captured by a time-dependent dimensionless multiplier \({Y}_{\theta }(t)\) of the rate constant \(\beta\). This rate constant, which appears in Appendix Eqs. 1–4, 18–22, and 24, determines the rate of disease transmission within the subpopulation unprotected by social-distancing behaviors when \({Y}_{\theta }\left(t\right)=1\). We take \({Y}_{\theta }\left(t\right)=1\) for times \(t<{\theta }_{1}\), i.e., for the initial period of the COVID-19 pandemic in the US that we take to have started on January 21, 2020. We take \({Y}_{\theta }\left(t\right)\) to have the form of a step function with distinct values greater than 1 for periods starting at \(t={\theta }_{1}\) and \({\theta }_{k+1}>{\theta }_{k}\) for \(k=1,\dots ,m.\) Thus, the model allows for \(m\) distinct periods of variant strain dominance delimited by a set of start times \(\theta =\{{\theta }_{1},\dots ,{\theta }_{m}\}\). We considered \(m=2\). We assume that variants differ only in transmissibility (Color Figure Online)
In these equations, the independent variable is time \(t\), and the state variables (\({S}_{M}\), \({S}_{P}\), \({E}_{1,M},\dots ,{E}_{5,M}\), \({E}_{1,P},\dots ,{E}_{5,P}\), \({E}_{2,Q},\dots ,{E}_{5,Q}\), \({A}_{M}\), \({A}_{P}\), \({A}_{Q}\), \({I}_{M}\), \({I}_{P}\), \({I}_{Q}, {H}_{U}\), \(D\), \({R}_{U}\), \({V}_{1},\dots , {V}_{6}\), \({S}_{V,1},\dots , {S}_{V,4}\), \({E}_{V}\), \({A}_{V}\), \({I}_{V}\), \({H}_{V}\), and \({R}_{V}\)) represent 40 (sub)populations (Table 5 in Appendix), which change over time. Thus, each ODE in Eqs. (1)–(28) defines the time-rate of change of a population, i.e., the time-rate of change of a state variable. Note that Eqs. (5), (6), (8) and (19) define 4, 4, 3, and 5 ODEs of the model, respectively. The model is formulated such that \({S}_{0}\), the total population, is a constant. Thus, the model does not account for birth, death for reasons other than COVID-19, immigration, or emigration.
The initial condition associated with Eqs. (1)–(28) is taken to be \({S}_{M}\left({t}_{0}\right)={S}_{0}\), \({I}_{M}\left({t}_{0}\right)={I}_{0}=1\), and all other populations (\({S}_{P}\), \({E}_{1,M},\dots ,{E}_{5,M}\), \({E}_{1,P},\dots ,{E}_{5,P}\), \({E}_{2,Q},\dots ,{E}_{5,Q}\), \({A}_{M}\), \({A}_{P}\), \({A}_{Q}\), \({I}_{P}\), \({I}_{Q}, {H}_{U}\), \(D\), \({R}_{U}\), \({V}_{1},\dots , {V}_{6}\), \({S}_{V,1},\dots , {S}_{V,4}\), \({E}_{V}\), \({A}_{V}\), \({I}_{V}\), \({H}_{V}\), and \({R}_{V}\)) are equal to 0. Recall that the parameter \({S}_{0}\) denotes the total region-specific population size. Thus, we assume that the entire population is susceptible at the start of the local epidemic at time \(t={t}_{0}>0\), where time \(t=0\) corresponds to 00:00 h (midnight) on January 21, 2020. The parameter \({I}_{0}\) denotes the number of infectious symptomatic persons at the start of the regional epidemic.
In the model, the parameters \(\beta\), \({k}_{L}\), \({k}_{Q}\), \({j}_{Q}\), \({c}_{A}\), \({c}_{I}\), \({c}_{H}\), and \({k}_{V}\) are positive-valued rate constants (all with units of \({\text{d}}^{-1}\)), and the parameters \({m}_{b}\), \({m}_{h}\), \({f}_{A}\), \({f}_{H}\), \({f}_{R}\), \({f}_{0}\ge {f}_{1}\), \({f}_{1}\ge {f}_{2}\), and \({f}_{2}\) are (dimensionless) fractions. Brief definitions of parameters are given in Tables 1 and 2.
In the model, the quantities \({\phi }_{M}(t,\rho )\), \({\phi }_{P}(t,\rho )\), and \({\phi }_{V}\left(t\right)\) are functions of (time-dependent) state variables (as defined below), which represent the population of infectious persons who are mixing freely (i.e., not practicing social-distancing), the population of infectious persons who are practicing social-distancing (i.e., adopting disease-avoiding behaviors), and the population of persons eligible for vaccination, respectively. The quantities \({\phi }_{M}(t,\rho )\) and \({\phi }_{P}(t,\rho )\) are also functions of \(\rho \equiv ({\rho }_{E},{\rho }_{A})\), where \({\rho }_{E}\) (\({\rho }_{A}\)) is a dimensionless ratio representing the infectiousness of persons in the incubation phase of infection (the infectiousness of asymptomatic persons in the immune clearance phase of infection) relative to the infectiousness of symptomatic persons with the same social-distancing behavior. The quantity \({\phi }_{V}(t)\) represents the population of persons eligible for vaccination.
The state variables that appear in these equations represent time-varying populations. Recall that state variables are defined in Table 5 in Appendix.
In the model, the quantities \({U}_{\sigma }(t)\), \({U}_{{\theta }_{1}}(t)\), and \({U}_{{\theta }_{2}}(t)\) are unit step functions. The values of these functions change from 0 to 1 at the times indicated by the subscripts: \(\sigma\), the onset time of the initial social-distancing period; \({\theta }_{1}\), the takeoff time of SARS-CoV-2 variant Alpha; and \({\theta }_{2}\), the takeoff time of SARS-CoV-2 variant Delta.
As indicated in Fig. 7 in Appendix, transitions from \({S}_{M}\) to \({S}_{P}\), for example, become possible at time \(t=\sigma\), transitions from \({S}_{V,2}\) to \({E}_{V}\) become possible at time \(t={\theta }_{1}\), and transitions from \({S}_{V,3}\) to \({E}_{V}\) become possible at time \(t={\theta }_{2}\).
In the model, the quantities \({P}_{\tau }\left(t\right)\), and \({\Lambda }_{\tau }(t)\) are step functions that characterize changes in social-distancing. The value of \({P}_{\tau }(t)\) determines a setpoint steady-state fraction of susceptible persons who are practicing social-distancing. The value of \({\Lambda }_{\tau }\left(t\right)\) determines a time scale for approach to the setpoint steady state. Changes in the values of \({P}_{\tau }(t)\) and \({\Lambda }_{\tau }(t)\) occur coordinately. These changes occur at times \(t=\sigma ,{\tau }_{1},\dots , {\tau }_{n}\), where \(n\) is the number of distinct social-distancing periods beyond an initial social-distancing period. Initially, we took \(n=7\) (i.e., 8 total social-distancing stages). The value of \(n\) is decremented by 1 (at an inferred time) if \(n\leftarrow n-1\) is determined to be admissible by a model-selection procedure, which is described below. It should be noted that \({{p}_{0}, p}_{1},\dots ,{p}_{n}\) are parameters of \({P}_{\tau }(t)\) and that \({{\lambda }_{0}, \lambda }_{1},\dots ,{\lambda }_{n}\) are parameters of \({\Lambda }_{\tau }(t)\). These parameters determine the non-zero values of the step functions over different periods. For example, \({p}_{1}\) is the value of \({P}_{\tau }(t)\) and \({\lambda }_{1}\) is the value of \({\Lambda }_{\tau }(t)\) for the period \(t\in [{\tau }_{1},{\tau }_{2})\).
The values of \({P}_{\tau }(t)\) and \({\Lambda }_{\tau }(t)\) are 0 for \(t<\sigma\).
In the model, the quantity \(\mu (t)\) is a piecewise linear interpolant to a function \(\widetilde{\mu }\left(t\right)\) that characterizes the current rate of vaccination. The value of \(\widetilde{\mu }\left(t\right)\) is determined by the empirical daily rate of vaccination, and thus, can vary from day to day. Daily vaccination data were extracted from the Covid Act Now database and the Democrat and Chronicle newspaper (footnote 8) using the Covid Act Now Data API (footnote 7) and web scraping. We will use \({\mu }_{i}\) to refer to the value of \(\widetilde{\mu }\left(t\right)\) for \(t\in \left[{t}_{i},{t}_{i+1}\right)\), where time \({t}_{i}\) corresponds to midnight on the \(i\)th day after January 21, 2020.
Settings for \({\mu }_{i}\) were made such that \({\mu }_{i}{S}_{0}\times 1 {\text{d}}\) is the number of vaccinations completed in the nearest past 1-d period according to Covid Act Now data and the Democrat and Chronicle newspaper.
In the model, the quantity \({Y}_{\theta }(t)\) is a step function that quantifies how disease transmissibility increases upon emergence of SARS-CoV-2 variants Alpha and Delta. Initially, \({Y}_{\theta }\left(t\right)=1\). The value of \({Y}_{\theta }(t)\) is increased (by an inferred factor greater than 1 at an inferred time, \({\theta }_{1}\) or \({\theta }_{2}\)) if the change is determined to be admissible by a model-selection procedure, which is described below. It should be noted that \({y}_{1}\) and \({y}_{2}\) are parameters of \({Y}_{\theta }(t)\). These parameters determine the values of the step function \({Y}_{\theta }(t)\) over different periods: \({y}_{1}\) is the value of \({Y}_{\theta }(t)\) for the period \(t\in [{\theta }_{1},{\theta }_{2})\) and \({y}_{2}\) is the value of \({Y}_{\theta }(t)\) for the remaining period of concern (with Delta as the dominant circulating viral strain).
where \(m\) is the number of viral variants that have emerged up to the current time, \({\theta }_{0}\equiv {t}_{0}\), and \({y}_{0}\equiv 1\). Here, we consider \(m= 2\).
1.3 Equations of the Auxiliary Measurement Model
As in the study of Lin et al. (2021), we assumed that only symptomatic persons are detected in testing. The accumulation of symptomatic persons is governed by
where \({C}_{S}(t)\) is the cumulative number of symptomatic persons (cases) at time \(t\). Here, unlike in the study of Lin et al. (2021), the expression for \({C}_{S}(t)\) accounts for exposed persons in quarantine. Initially, \({C}_{S}=0.\) We numerically integrated Appendix Eq. (39) together with the ODEs of the compartmental model. From the trajectory for \({C}_{S}\), we derive a prediction for the expected number of new COVID-19 cases reported on calendar date \({\mathcal{D}}_{{\text{i}}}\), \(I\left({t}_{i},{t}_{i+1}\right)\), using the following equation:
where \({f}_{D}\) is an adjustable region-specific parameter characterizing the time-averaged fraction of symptomatic cases detected and reported, \({t}_{i}\) corresponds to midnight on the \(i\)th day after January 21, 2020, and \({t}_{i+1}-{t}_{i}\) is the reporting interval (1 d). We compare \(I({t}_{i},{t}_{i+1})\) to \(\delta {C}_{i}\), the number of new cases reported on calendar date \({\mathcal{D}}_{{\text{i}}}\).
1.4 Definition of the Likelihood Function
Bayesian inference relies on the definition of a likelihood, which here serves the purpose of assessing the compatibility of available surveillance data with adjustable (free) parameter values. Let us use \({\left\{\delta {C}_{i}\right\}}_{i=0}^{d}\) to denote the daily case reporting data available between 0 and \(d\) days after midnight on January 21, 2020 (the date of the first case report in the US) and let \({D=\left\{\delta {C}_{i}\right\}}_{i=0}^{d}\). Let us use \({\theta }_{F}(n,m)\) to denote the set of adjustable (free) parameter values. The number of adjustable parameters, \(|{\theta }_{F}|\), depends on \(n\), the number of social-distancing periods considered beyond an initial social-distancing period, and \(m\), the number of SARS-CoV-2 variants under consideration. As in the study of Lin et al. (2021), we assume that \(\delta {C}_{i}\), the number of new COVID-19 cases detected over a 1-d period and reported on calendar date \({\mathcal{D}}_{{\text{i}}}\) for a given region, is a random variable and its expected value follows a model-derived deterministic trajectory given by \(I({t}_{i},{t}_{i+1})\) (Eq. 40). We assume that day-to-day fluctuations in the random variable are independent and characterized by a negative binomial distribution \({\text{NB}}\left(r,{q}_{i}\right)\), which has two parameters, \(r>0\) and \({q}_{i}\in (\mathrm{0,1})\). Note that \({\mathbb{E}}\left[{\text{NB}}(r,{q}_{i})\right]=r(1-{q}_{i})/{q}_{i}\). We assume that this distribution has the same dispersion parameter \(r\) across all case reports. With these assumptions, we arrive at the following likelihood function:
where
and
In these equations, \(i\) is an integer indicating the date \({\mathcal{D}}_{i}\) and period \(\left({t}_{i},{t}_{i+1}\right)\); \({\text{nbinom}}\left(\delta {C}_{i};r,{q}_{i}\right)\) is the probability mass function of the negative binomial distribution \({\text{NB}}(r,{q}_{i})\), and \({\theta }_{F}(n,m)=\{{t}_{0},\beta , \sigma , {\tau }_{1},\dots ,{\tau }_{n},{p}_{0},{p}_{1,}\dots ,{p}_{n},{\lambda }_{0},{\lambda }_{1},\dots ,{\lambda }_{n},{\theta }_{1},\dots ,{\theta }_{m}, {y}_{1},\dots ,{y}_{m},{f}_{D}, r\}\) for \(n\ge 1\) and \(m\ge 1\); \({\theta }_{F}(\mathrm{0,0})=\{{t}_{0},\beta ,{p}_{0},{\lambda }_{0},{f}_{D}, r\}\).
1.5 Parameters
Each model parameter is briefly described in Tables 1, 2, and 3. These parameters have either fixed values or adjustable values (i.e., values inferred from surveillance data). The fixed values may be universal (i.e., applicable to all MSAs of interest) or MSA-specific. All inferred parameter values are MSA-specific. In addition, the measurement model (Appendix Eqs. 39 and 40) has one adjustable MSA-specific parameter, \({f}_{D}\), and the likelihood function (Appendix Eqs. 41–43) has one adjustable MSA-specific parameter, \(r\). Values of the other likelihood parameters, \({q}_{0},\dots ,{q}_{d}\), are constrained and are determined using Appendix Equation 43.
1.6 Original Model of Lin et al.
The model shares \(19+3n\) parameters with the model of Lin et al. (2021), including parameters that define the initial condition (\({t}_{0}\), \({I}_{0}\), and \({S}_{0}\)). (Recall that \(n\) is the number of social-distancing periods being considered beyond the initial social-distancing period.) The shared parameters are \({t}_{0}\), \({I}_{0}\), \({S}_{0}\), \(\beta\), \(\sigma\), \({\tau }_{1},\dots , {\tau }_{n}\), \({p}_{0},\dots ,{p}_{n}\), \({\lambda }_{0},\dots ,{\lambda }_{n}\), \({\rho }_{A}\), \({\rho }_{E}\), \({m}_{b}\), \({f}_{A}\), \({f}_{H}\), \({f}_{R}\), \({k}_{L}\), \({k}_{Q}\), \({j}_{Q}\), \({c}_{A}\), \({c}_{H}\), and \({c}_{I}\). As in the study of Lin et al. (2021), we inferred MSA-specific values for the following parameters: \({t}_{0}\), \(\beta\), \(\sigma\), \({p}_{0},\dots , {p}_{n}\), and \({\lambda }_{0},\dots , {\lambda }_{n}\). We also inferred MSA-specific values for \({\tau }_{1},\dots ,{\tau }_{n}\) provided that \(n\ge 1\). As in the study of Lin et al. (2021), the remaining 14 parameters shared between the old and new models (\({I}_{0}\), \({S}_{0}\), \({\rho }_{A}\), \({\rho }_{E}\), \({m}_{b}\), \({f}_{A}\), \({f}_{H}\), \({f}_{R}\), \({k}_{L}\), \({k}_{Q}\), \({j}_{Q}\), \({c}_{A}\), \({c}_{H}\), and \({c}_{I}\)) were taken to have fixed values, and we adopted the settings of Lin et al. (2021) for these parameters (Table 3). These settings are universal except for the setting for \({S}_{0}\), the total population, which is MSA-specific.
1.7 Extension of the Model of Lin et al.
Our extension of the model of Lin et al. (2021) introduces \(5+2\left(m+1\right)+(d+1)\) parameters, where \(m\) (\(=0\), 1 or 2) is the number of SARS-CoV-2 variants being considered and \(d\) is the number of days since January 21, 2020: \({\theta }_{0}, \dots ,{\theta }_{m}\), \({y}_{0},\dots ,{y}_{m}\), \({m}_{h}\), \({f}_{0}\), \({f}_{1}\), \({f}_{2}\), \({k}_{V}\), and \({\mu }_{0},\dots ,{\mu }_{d}\). The \(\theta\) and \(y\) parameters are variant takeover times and transmissibility factors, respectively, except that the value of \({\theta }_{0}\) is defined as \({t}_{0}\) and the value of \({y}_{0}\) is defined as 1. The Alpha transmissibility factor \({y}_{1}\), the Alpha takeoff time \({\theta }_{1}\), the Delta transmissibility factor \({y}_{2}\), and the Delta takeoff time \({\theta }_{2}\) were inferred for each MSA with \(m=2\) (cf. Table 1). The transmissibility factors were each constrained to be greater than or equal to 1. The settings for \({\mu }_{0},\dots ,{\mu }_{d}\) are empirical and MSA-specific. Each \({\mu }_{i}\) is set such that \({\mu }_{i}{S}_{0}\times 1 {\text{d}}\) is the number of vaccinations completed over the past 1-d period nearest to the \(i\)th day after January 21, 2020. As noted earlier, the number of completed vaccinations was obtained for each MSA from Covid Act Now and the Democrat and Chronicle newspaper (footnote 8) using the Covid Act Now Data API (footnote 7) and web scraping. In the spreadsheet accessed daily from Covid Act Now, the ‘metrics.vaccinationsCompletedRatio’ column gives the percentage of the total population that has received the recommended number of doses: one dose for Ad26.CoV2.S (Janssen, Johnson & Johnson) or two doses for mRNA-1273 (Moderna) and BNT162b2 (Pfizer-BioNTech). As a simplification, we considered all completed vaccinations to be equivalent. The parameters \({m}_{h}\), \({f}_{0}\), \({f}_{1}\), \({f}_{2}\), and \({k}_{V}\) were assigned fixed universal estimates (Table 3). Each of these estimates is explained below.
1.8 Estimation of \({{{k}}}_{{{V}}}\)
The rate constant \({k}_{V}\) characterizes the rate of transition out of compartment \({V}_{i}\) for \(i=1,\dots ,{n}_{V}\). Recall that, in the model, susceptible persons enter \({V}_{1}\) upon vaccination (Fig. 1, Fig. 7 in Appendix). The values of \({n}_{V}\) (\(=6\)) and \({k}_{V}\) (\(=0.3 \, {\text{d}}^{-1}\)) were selected so that the time a person spends in \({V}_{1},\dots ,{V}_{{n}_{V}}\), which we will denote as \({t}_{V}\), is distributed approximately the same as \({\widetilde{t}}_{V}\), the waiting time between vaccination of a previously uninfected person and detection of vaccine-induced SARS-CoV-2-specific IgG antibodies (Korodi et al. 2022) (Fig. 8 in Appendix). According to the model, the time that a person spends in \({V}_{1},\dots ,{V}_{{n}_{V}}\) is distributed according to the probability density function \(f\left({t}_{V};{n}_{V},{k}_{V}\right)={k}_{V}^{{n}_{V}}{t}_{V}^{{n}_{V}-1}{e}^{-{k}_{V}{t}_{V}}/\left({n}_{V}-1\right)!\), i.e., \({t}_{V}\) is Erlang distributed with shape parameter \({n}_{V}=6\) and rate parameter \({k}_{V}=0.3\) \({\text{d}}^{-1}\). As can be seen in Fig. 8 in Appendix, the cumulative distribution function of this Erlang distribution reasonably captures the empirical cumulative distribution of waiting times observed in the longitudinal study of Korodi et al. (2022). Thus, in the model, passage through \({V}_{1},\dots ,{V}_{6}\) with rate constant \({k}_{V}=0.3 \, {\text{d}}^{-1}\) accounts for the variable and significantly non-zero amount of time required for development of a protective antibody response after vaccination.
Comparison of an Erlang cumulative distribution function with shape parameter \({n}_{V}=6\) and rate parameter \({k}_{V}=0.3 {\text{d}}^{-1}\) and the empirical cumulative distribution of waiting times (\({\widetilde{t}}_{V}\) values) observed in the longitudinal study of Korodi et al. (Bo et al. 2021). The waiting time \({\widetilde{t}}_{V}\) is the time between vaccination of a previously uninfected person and detection of vaccine-induced SARS-CoV-2-specific IgG antibodies (color figure online)
1.9 Estimation of \({{{f}}}_{0}, {{{f}}}_{1},\) and \({{{f}}}_{2}\)
The parameters \({f}_{0}>{f}_{1}\), \({f}_{1}>{f}_{2}\), and \({f}_{2}\) are fractions that characterize the average effectiveness of vaccines used in the US and that determine the sizes of (mutually exclusive) subpopulations of vaccinated persons having different susceptibilities to productive infection (i.e., an infection that can be transmitted to others): \({S}_{V,1}\), \({S}_{V,2}\), \({S}_{V,3}\), and \({S}_{V,4}\) (Fig. 1, Fig. 7 in Appendix). We assume that persons in the \({S}_{V,1}\) subpopulation are susceptible to productive infection by any of the viral strains considered, and in contrast, we assume that persons in the \({S}_{V,4}\) subpopulation are susceptible to productive infection by none of the viral strains considered. Persons in the \({S}_{V,2}\) subpopulation are taken to be susceptible to productive infection by the Alpha and Delta variants but not viral strains in circulation before the emergence of Alpha. Persons in the \({S}_{V,3}\) subpopulation are taken to be susceptible to productive infection by the Delta variant but not the Alpha variant or viral strains in circulation before the emergence of Alpha. The quantity \(1-{f}_{0}\) defines the fraction of vaccinated persons who enter the \({S}_{V,1}\) subpopulation after exiting \({V}_{6}\), the quantity \({f}_{0}-{f}_{1}\) defines the fraction of vaccinated persons who enter the \({S}_{V,2}\) subpopulation after exiting \({V}_{6}\), the quantity \({f}_{1}-{f}_{2}\) defines the fraction of vaccinated persons who enter the \({S}_{V,3}\) subpopulation after exiting \({V}_{6}\), and \({f}_{2}\) defines the fraction of vaccinated persons who enter the \({S}_{V,4}\) subpopulation after exiting \({V}_{6}\). We take \({f}_{0}\) to characterize vaccine effectiveness before the emergence of Alpha. According to Thompson et al. (Thompson et al. 2021), vaccine effectiveness was initially 90%. Thus, we set \({f}_{0}=0.9\). We take \({f}_{1}\) to characterize vaccine effectiveness after the emergence of Alpha but before the emergence of Delta. According to Puranik et al. (2024), in May 2021, vaccine effectiveness was 81%. Thus, we set \({f}_{1}=0.81\). We take \({f}_{2}\) to characterize vaccine effectiveness after the emergence of Delta. According to Tang et al. (2021), the effectiveness of two doses of the Pfizer-BioNTech vaccine (BNT162b2) against Delta is 53.5% and the effectiveness of two doses of the Moderna vaccine (mRNA-1273) against Delta is 84.8%. Taking the average of these figures, we set \({f}_{2}=0.69\).
1.10 Estimation of \({{{m}}}_{{{h}}}\)
The parameter \({m}_{h}\) characterizes the reduced risk of severe disease for a vaccinated person in the case of a breakthrough infection. We set \({m}_{h}=0.04\), i.e., we assumed that there is a 25-fold reduction in the risk of severe disease for infected persons who have been vaccinated, which is consistent with the observations of Lopez Bernal et al. (2021).
1.11 Notable New Modeling Assumptions
It should be noted that we treat the incubation period for newly infected (exposed) vaccinated persons differently than for newly infected (exposed) unvaccinated persons (Fig. 1, Fig. 7 in Appendix). For unvaccinated persons, as in the study of Lin et al. (2021), we divide exposed persons in the incubation period of infection into five subpopulations: \({E}_{1,M},\dots ,{E}_{5,M}\) for exposed persons who are mixing (i.e., persons who are not practicing social-distancing), \({E}_{1,P},\dots , {E}_{5,P}\) for exposed persons who are practicing social-distancing, and \({E}_{1,Q},\dots , {E}_{5,Q}\) for exposed quarantined persons. Persons move through the five stages of the incubation period sequentially. In contrast, as a simplification, for vaccinated persons, we consider only a single exposed population: \({E}_{V}\). We take persons to exit \({E}_{V}\) with rate constant \({k}_{L}/5\) (Fig. 7 in Appendix). With this choice, the duration of the incubation period of infection is the same, on average, for both vaccinated and unvaccinated persons. The average duration is \(5/{k}_{L}\) (about 5 d) in both cases. The difference is that the duration of the incubation period is Erlang distributed for unvaccinated persons, as discussed by Lin et al. (2021), but exponentially distributed for vaccinated persons.
As indicated in Eq. (29), we take vaccinated persons with productive infections to be equally as infectious as unvaccinated persons.
As noted earlier, we take all vaccinated persons to be mixing (i.e., to not be practicing social-distancing). Thus, populations of infected vaccinated persons (\({E}_{V}\), \({I}_{V}\), and \({A}_{V}\)) contribute to \({\phi }_{M}(t)\) (Appendix Eq. 29) but not \({\phi }_{P}\left(t\right)\) (Appendix Eq. 30).
As indicated in Appendix Eq. 31, we consider pre-symptomatic exposed and asymptomatic unvaccinated persons to be eligible for vaccination and, thus, these persons contribute to the consumption of vaccine doses (i.e., these persons account for a portion of the number of completed vaccinations on a given day \(i\), \({\mu }_{i}{S}_{0}\times 1 {\text{d}}\)). However, we do not move these persons to vaccinated compartments. The reason is that exposed and asymptomatic persons are expected to develop immunity faster through recovery from infection (i.e., movement to \({R}_{U}\)) than from vaccination.
As indicated in Appendix Eq. 31, we do not consider symptomatic, quarantined, severely ill and hospitalized/isolated-at-home, and deceased persons to be eligible for vaccination.
1.12 Inference Approach
Recall that \({\theta }_{F}\) denotes the set of all adjustable parameters. As in the study of Lin et al. (2021), for each MSA, we inferred MSA-specific adjustable parameter values \({\theta }_{F}\) using all MSA-specific surveillance data available up to a specified day of inference \({\mathcal{D}}_{{\text{d}}}\) (i.e., the \(d\)th day after January 21, 2020). We took a Bayesian inference approach, meaning that, for a given dataset, we generated parameter posterior samples (a collection of \({\theta }_{F}\)’s) through Markov chain Monte Carlo (MCMC) sampling. The parameter posterior samples provide a probabilistic characterization of the adjustable parameter values consistent with the dataset used in inference. By drawing samples from the parametric posterior distribution, we generated a posterior predictive distribution for \(I({t}_{i},{t}_{i+1})\) for each \(i\) of interest. We considered all days from January 21, 2020 to October 31, 2021. In other words, for each \(i\) of interest, a prediction for \(I({t}_{i},{t}_{i+1})\) was generated for each of many \({\theta }_{F}\)’s drawn randomly (with uniform probability) from the parametric posterior distribution. The resulting distribution of \(I({t}_{i},{t}_{i+1})\) values is the posterior predictive distribution for \(I({t}_{i},{t}_{i+1})\). Recall that \(I({t}_{i},{t}_{i+1})\) is given by Appendix Eq. 40 and corresponds to \({\mathbb{E}}\left[\delta {C}_{i}\right]\), the expected number of new COVID-19 cases detected over a 1-d surveillance interval and reported for the \(i\)th day after January 21, 2020. Observation noise was injected into the posterior predictive distributions by replacing each sampled value for \(I({t}_{i},{t}_{i+1})\) with \({X}_{i}\sim {\text{NB}}(r,{q}_{i})\), where \(r\) is a member of the sampled set of parameter values \({\theta }_{F}\) used to generate the prediction of \(I({t}_{i},{t}_{i+1})\) and \({q}_{i}\) is given by Eq. 43.
According to Bayes’ theorem, given surveillance data \(D={\left\{\delta {C}_{i}\right\}}_{i=0}^{d}\), the parametric posterior is given as
where \({\mathbb{P}}\left\{{\theta }_{F}\right\}\) is the prior (which is formulated to capture knowledge of \({\theta }_{F}\) external to \(D\) or to express lack of such knowledge), \({\mathbb{P}}\left\{D|{\theta }_{F}\right\}\) is the likelihood defined by Appendix Eqs. 41–43, and \(Z\) is a normalizing constant. We assumed a proper uniform prior, i.e., for each adjustable parameter, we assumed that all values between specified lower and upper bounds are equally likely before consideration of \(D\). We used the same bounds as in the study of Lin et al. (2021). Then, we used an adaptive MCMC (aMCMC) algorithm (Andrieu and Thoms 2008) to generate samples from \({\mathbb{P}}\left\{D|{\theta }_{F}\right\}{\mathbb{P}}\left\{{\theta }_{F}\right\}\), which is proportional to \({\mathbb{P}}\left\{{\theta }_{F}|D\right\}\). Thus, the relative probabilities of parameter sets \({\theta }_{F}\) according to \({\mathbb{P}}\left\{{\theta }_{F}|D\right\}\) are correctly represented by the samples.
Specifically, the adaptive MCMC algorithm (Andrieu and Thoms 2008) generates samples from the multivariate parametric posterior for adjustable model parameters (\({t}_{0}, \beta\), and parameters for variant emergence and social-distancing), the measurement model parameter \({f}_{D}\), and the likelihood parameter \(r\) (Tables 1 and 2). This algorithm is available within the PyBioNetFit software package (Neumann et al. 2022). Use of the algorithm was performed as described by Lin et al. (2021). The report of Neumann et al. (2022) includes helpful general usage advice, which was followed in this study. Inference jobs were executed on a computer cluster. For each inference job, a total of 25 chains were generated, and the chain with the best mixing and convergence properties was selected for subsequent analyses.
Each inference was conditioned on the compartmental model of Fig. 1 (Appendix Eqs. 1–38), settings for the structural parameters \(m\) (the number of SARS-CoV-2 variants under consideration) and \(n\) (the number of social-distancing periods under consideration beyond an initial social-distancing period), the measurement model (Appendix Eqs. 39 and 40), and fixed parameter estimates (Tables 1 and 2), including empirical daily per capita vaccination rates (i.e., the settings for \({\mu }_{i}\) in Appendix Eq. 37). We assumed a proper uniform prior for each adjustable parameter (Lin et al. 2021) and a negative binomial likelihood function (Appendix Eqs. 41–43). Use of proper uniform priors means that MAP estimates are maximum likelihood estimates (MLEs). In each inference, the data entering the likelihood function, \(D={\left\{\delta {C}_{i}\right\}}_{i=0}^{d}\) (Appendix Eq. 41), were MSA-specific daily reports of newly detected COVID-19 cases available up to the date of inference \({\mathcal{D}}_{{\text{d}}}\) (i.e., the \(d\)-th day after January 21, 2020). Thus, all inferences are region-specific and time-dependent.
1.13 Use of Model Selection to Determine Intervals of Step Functions
Variant takeover times, \(\theta =({\theta }_{1},{\theta }_{2})\), and start times of social-distancing periods, \(\tau =({\sigma , \tau }_{1},\dots , {\tau }_{n})\), were inferred from data; however, changes of the associated time-dependent step functions, \({Y}_{\theta }\left(t\right)\), \({P}_{\tau }(t)\), and \({\Lambda }_{\tau }(t)\), were introduced only when an increase in model complexity was deemed to be justified. Each decision to introduce variant takeover or start of a new social-distancing period (beyond the initial period) was made using a model-selection procedure, which is described below. It should be noted that \({y}_{1}\) and \({y}_{2}\) are parameters of \({Y}_{\theta }(t)\), \({{p}_{0},p}_{1},\dots ,{p}_{n}\) are parameters of \({P}_{\tau }(t)\), and \({{\lambda }_{0},\lambda }_{1},\dots ,{\lambda }_{n}\) are parameters of \({\Lambda }_{\tau }(t)\). These parameters determine the values of the step functions over different periods. For example, for \(n\ge 1\), \({p}_{1}\) is the value of \({P}_{\tau }(t)\) and \({\lambda }_{1}\) is the value of \({\Lambda }_{\tau }(t)\) for the period \(t\in [{\tau }_{1},{\tau }_{2})\). Similarly, \({y}_{1}\) is the value of \({Y}_{\theta }(t)\) for the period \(t\in [{\theta }_{1},{\theta }_{2})\), and \({y}_{2}\) is the value of \({Y}_{\theta }(t)\) for the period \(t\in [{\theta }_{2},{t}_{\text{final}})\), where \({t}_{\text{final}}\) corresponds to the date of inference (October 31, 2021).
We started with a setting of \(n=7\) for each MSA of interest (i.e., 8 total social-distancing stages). To determine if \(n\) could be reduced, we conducted parsimony checks. In a parsimony check, 100 MLE curves, each constituting a fit to the data, were generated via optimization jobs. In these jobs, the total number of social-distancing stages in the model (\(n+1\)) was set at 1 less than for the current proposed best fit. Each of the 100 fits was visually inspected to determine whether or not the following criteria held: (1) the quality of fit is acceptable (i.e., comparable to what is obtained with the current proposed best fit); (2) Alpha and Delta surges (identified by sequencing data) are explained at least partly by increased transmissibility; (3) social-distancing setpoint parameter values are feasible (i.e., each setpoint parameter takes on a value between 0.2 and 0.8); and (4) social-distancing changes proximal to an Alpha or Delta surge (if any) occur only after an increase in transmissibility. If one or more of these conditions was not satisfied, we accepted the proposed best fit as the most parsimonious fit to the data. If all conditions held, we updated the proposed best fit, and the parsimony check was repeated for a model with one less social-distancing stage. In the Appendix, we show examples of violations of parsimony-check criteria for the MSA surrounding Houston (Fig. 9 in Appendix).
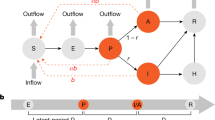
Example of a parsimony check with criteria that are violated for the MSA surrounding Houston. A Alpha and Delta surges (identified by sequencing data) are not explained, at least in part, by increased transmissibility, while all other criteria are satisfied. Each broken vertical black line from left to right indicates the date of onset of a social-distancing stage (i.e., with n = 3 for this illustration). Each broken vertical red line from left to right indicates the takeoff date of a variant (namely, Alpha or Delta). B Not all social-distancing setpoint parameter values are feasible, while all other criteria are satisfied. C Social-distancing changes proximal to an Alpha or Delta surge (if any) precede an increase in transmissibility, while all other criteria are satisfied. In Panel A, we see that the orange segment between the two broken vertical red lines, which denotes the relative transmissibility of Alpha, is no larger than the orange segment to its left, which denotes the relative transmissibility of ancestral strains of SARS-CoV-2. In Panel B, the rightmost blue segment, which denotes the social-distancing setpoint parameter value for the final social-distancing stage, has a value of roughly 0.16, which is deemed infeasible by our criterion. Finally, in Panel C, the rightmost broken vertical black line is proximal to the leftmost broken vertical red line, which corresponds to the takeoff time of the Alpha surge, and the social-distancing setpoint parameter value changes prior to the Alpha surge. Note that all three panels show MLE curves, each of which constitutes an acceptable fit to the data (Color Figure Online)
1.14 Simulations
After specification of parameter values (Tables 1, 2, and 3), we used the SciPy (https://www.scipy.org) interface to LSODA (Hindmarsh and Stepleman 1983) to numerically integrate the system of coupled ODEs consisting of the 40 ODEs of the compartmental model and the 1 ODE of the measurement model (Appendix Eqs. 1–39). The initial condition was defined by settings for \({t}_{0}\), \({I}_{0}\), and \({S}_{0}\) (Tables 1, 2, 3). Integration combined with use of Appendix Eq. 40 yielded a prediction of the expected number of new cases detected for each 1-d surveillance period of interest in the past or future: \(I({t}_{i},{t}_{i+1})\), where \({t}_{i}\) corresponds to midnight on the \(i\)th day after January 21, 2020. To account for randomness in case detection and reporting, we replaced \(I({t}_{i},{t}_{i+1})\) with \({X}_{i}\sim {\text{NB}}(r,{q}_{i})\), where \({q}_{i}\) is given by Appendix Eq. 43.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mallela, A., Chen, Y., Lin, Y.T. et al. Impacts of Vaccination and Severe Acute Respiratory Syndrome Coronavirus 2 Variants Alpha and Delta on Coronavirus Disease 2019 Transmission Dynamics in Four Metropolitan Areas of the United States. Bull Math Biol 86, 31 (2024). https://doi.org/10.1007/s11538-024-01258-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-024-01258-4