Abstract
Physiologically-based pharmacokinetic (PBPK) modeling is important for studying drug delivery in the central nervous system, including determining antibody exposure, predicting chemical concentrations at target locations, and ensuring accurate dosages. The complexity of PBPK models, involving many variables and parameters, requires a consideration of parameter identifiability; i.e., which parameters can be uniquely determined from data for a specified set of concentrations. We introduce the use of a local sensitivity-based parameter subset selection algorithm in the context of a minimal PBPK (mPBPK) model of the brain for antibody therapeutics. This algorithm is augmented by verification techniques, based on response distributions and energy statistics, to provide a systematic and robust technique to determine identifiable parameter subsets in a PBPK model across a specified time domain of interest. The accuracy of our approach is evaluated for three key concentrations in the mPBPK model for plasma, brain interstitial fluid and brain cerebrospinal fluid. The determination of accurate identifiable parameter subsets is important for model reduction and uncertainty quantification for PBPK models.











Similar content being viewed by others
Availability of data and materials
Data sharing is not applicable to this article as no datasets were analyzed or generated during the current study.
References
Apgar JF, Witmer DK, White FM, Tidor B (2010) Sloppy models, parameter uncertainty, and the role of experimental design. Mol BioSyst 6:1890–1900
Banks HT, Bekele-Maxwell K, Bociu L, Noorman M, Tillman K (2015) The complex-step method for sensitivity analysis of non-smooth problems arising in biology. Eurasian J Math Comput Appl 3:15–68
Bloomingdale P, Bakshi S, Maass C, van Maanen E, Pichardo-Almarza C, Yadav DB, van der Graaf P, Mehrotra N (2021) Minimal brain PBPK model to support the preclinical and clinical development of antibody therapeutics for CNS diseases. J Pharmacokinet Pharmacodyn 48(6):861–871
Bellu G, Saccomani MP, Audoly S, D’Angiò L (2007) DAISY: a new software tool to test global identifiability of biological and physiological systems. Comput Methods Programs Biomed 88(1):52–61
Brown LV, Coles MC, McConnell M, Ratushny AV, Gaffney EA (2022) Analysis of cellular kinetic models suggest that physiologically based model parameters may be inherently, practically unidentifiable. J Pharmacokinet Pharmacodyn 49(5):539–556
Burth M, Verghese GC, Vélez-Reyes M (1999) Subset selection for improved parameter estimates in on-line identification of a synchronous generator. IEEE Trans Power Syst 14(1):218–225
Calvier EAM, Nguyen TT, Johnson TN, et al (2018) Can population modelling principles be used to identify key PBPK parameters for paediatric clearance predictions? An innovative application of optimal design theory. Pharm Res 209
Carter SJ, Chouhan B, Sharma P, Chappell MJ (2020) Prediction of clinical transporter-mediated drug-drug interactions via comeasurement of pitavastatin and eltrombopag in human hepatocyte models. CPT Pharmacometrics Syst Pharmacol 9(4):211–221
Chang HY, Wu S, Meno-Tetang G, Shah DK (2019) A translational platform PBPK model for antibody disposition in the brain. J Pharmacokinet Pharmacodyn 46(4):319–338
Cintrón-Arias A, Banks HT, Capaldi A, Lloyd AL (2009) A sensitivity matrix based methodology for inverse problem formulation. J Inverse Ill Posed Probl 17(6):545–564
Clewell HJ III, Lee TS, Carpenter RL (1994) Sensitivity of physiologically based pharmacokinetic models to variation in model parameters: methylene chloride. Risk Anal 1994(14):521–531
Dwek R (2009) Antibodies and antigens: it’s all about the numbers game. Proc Natl Acad Sci USA 106(7):2087–2088
Efron B, Tibshirani RJ (1993) An introduction to the bootstrap. Chapman & Hall/CRC, Boca Raton
Evans MV, Eklund CR, Williams DN, Sey YM, Simmons JE (2020) Global optimization of the Michaelis-Menten parameters using physiologicallybased pharmacokinetic (PBPK) modeling and chloroform vapor uptake data in F344 rats. Inhalation Toxicol 32(3):97–109
Evans MV, Andersen ME (1995) Sensitivity analysis and the design of gas uptake inhalation studies. Inhalation Toxicol 7(7):1075–1094
Friswell MI, Penny JET, Garvey SD (1997) Parameter subset selection in damage location. Inverse Probl Eng 5:189–215
Gautschi W (1997) Numerical analysis. Birkhäuser, Basel
Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP (2007) Universally sloppy parameter sensitivities in systems biology models. PLoS Comput Biol 3(10):1871–78
Hong H, Ovchinnikov A, Pogudin G, Yap C (2019) SIAN: software for structural identifiability analysis of ODE models. Bioinformatics 35(16):2873–2874
Hsieh NH, Reisfeld B, Bois FY, Chiu WA (2018) Applying a global sensitivity analysis workflow to improve the computational efficiencies in physiologically-based pharmacokinetic modeling. Front Pharmacol 9:588
Ipsen ICF (2009) Numerical matrix analysis. SIAM, Philadephia
Jones H, Rowland-Yeo K (2013) Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. CPT Pharmacometrics Syst Pharmacol 2(8):e63
Keene ON (1995) The log transformation is special. Stat Med 14(8):811–819
Kendrick F, Evans ND, Arnulf B, Avet-Loiseau H, Decaux O, Dejoie T, Fouquet G, Guidez S, Harel S, Hebraud B, Javaugue V, Richez V, Schraen S, Touzeau C, Moreau P, Leleu X, Harding S, Chappell MJ (2017) Analysis of a compartmental model of endogenous immunoglobulin G metabolism with application to multiple myeloma. Front Physiol 8:147
Kendrick F, Evans ND, Berlanga O, Harding SJ, Chappell MJ (2019) Parameter identification for a model of neonatal Fc receptor-mediated recycling of endogenous immunoglobulin G in humans. Front Immunol 10:674
Kim B, Lee JH (2019) Parameter subset selection and biased estimation for a class of ill-conditioned estimation problems. J Process Control 81:65–75
Koyama S, Toshimoto K, Lee W, Aoki Y, Sugiyama Y (2021) Revisiting nonlinear Bosentan pharmacokinetics by physiologically based pharmacokinetic modeling: target binding, albeit not a major contributor to nonlinearity, can offer prediction of target occupancy. Drug Metab Dispos 49(4):298–304
Lavezzi SM, Mezzalana E, Zamuner S, De Nicolao G, Ma P, Simeoni M (2018) MPBPK-TMDD models for mAbs: alternative models, comparison, and identifiability issues. J Pharmacokinet Pharmacodyn 45(6):787–802
Lyness J, Moler C (1967) Numerical differentiation of analytic functions. SIAM J Numer Anal 4:202–210
Martins JRRA, Sturdza P, Alonso JJ (2003) The complex-step derivative approximation. ACM Transactions on Mathematical Software 29(3):245–262
Matthews JL, Schultz IR, Easterling MR, Melnick RL (2009) Physiologically based pharmacokinetic modeling of dibromoacetic acid in F344 rats. Toxicol Appl Pharmacol 244(2010):196–207
Monsalve-Bravo GM, Lawson BAJ, Drovandi C, Burrage K, Brown KS, Baker CM, Vollert SA, Mengersen K, McDonald-Madden E, Adams MP (2022) Analysis of sloppiness in model simulations: Unveiling parameter uncertainty when mathematical models are fitted to data. Sci Adv 8(38):eabm595
National Human Genome Research Institute (2023). https://www.genome.gov/genetics-glossary/Antibody
Paul S (2011) Therapeutic antibodies for brain disorders. Sci Transl Med 3(84):1–5
Pearce KJ, Nellenbach K, Smith RC, Brown AC, Haider MA (2021) Modeling and parameter subset selection for fibrin polymerization kinetics with applications to wound healing. Bull Mathem Biol 83(5):1–22
Quaiser T, Mónnigmann M (2009) System identifiability testing for unambiguous mechanistic modeling—application to JAK-STAT, MAP kinase, and NF-\(\kappa \) B signaling pathway models. BMC Syst Biol 3(1):50
Raue A, Karlsson J, Saccomani MP, Jirstrand M, Timmer J (2014) Comparison of approaches for parameter identifiability analysis of biological systems. Bioinformatics 30(10):1440–1448
Slob W, Janssen PH, van den Hof JM (1997) Structural identifiability of PBPK models: practical consequences for modeling strategies and study designs. Crit Rev Toxicol 27(3):261–272
Smith RC (2014) Uncertainty quantification: theory, implementation, and applications. SIAM, Philadephia
Stigter JD, Molenaar J (2015) A fast algorithm to assess local structural identifiability. Automatica 58:118–124
Székely GJ, Rizzo ML (2004) Testing for equal distributions in high dimensions. InterStat 5:1249–1272
Székely GJ, Rizzo ML (2013) Energy statistics: a class of statistics based on distances. J Stat Plan Inference 143(8):1249–1272
Todd P, Brogden R, Muromonab C (1989) A review of its pharmacology and therapeutic potential. Drugs 37(6):871–899
Transtrum MK, Machta BB, Brown KS, Daniels BC, Myers CR, Sethna JP (2015) Perspective: sloppiness and emergent theories in physics, biology, and beyond. J Chem Phys 143(1):010901
Yates JW (2006) Structural identifiability of physiologically based pharmacokinetic models. J Pharmacokinet Pharmacodyn 33(4):421–439
Funding
This study was funded in part by grant DMS-1638521 from the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A
Appendix A
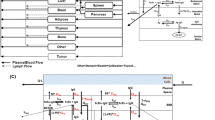
The brain minimal PBPK (mPBPK) model (Bloomingdale et al. (2021)) comprises the following system of ordinary differential equations for the 16 antibody concentrations:
-
1.
Plasma (\(C_P\))
$$\begin{aligned} V_{P} \cdot \frac{dC_P}{dt}= & {} (Q_T-L_T)\cdot C_{T_{V}}+(Q_B-L_B)\cdot C_{B_{V}}+(L_T+L_B)\cdot C_L\nonumber \\{} & {} -Q_T\cdot C_P-Q_B \cdot C_P \end{aligned}$$(15)Table 5 Parameter values for human in the model of Bloomingdale et al. 2021 -
2.
Tissue vascular (\(C_{T_{V}}\))
$$\begin{aligned} V_{T_{V}} \cdot \frac{dC_{T_{V}}}{dt}= & {} Q_T\cdot C_P-(Q_T-L_T)\cdot C_{T_{V}}-((1-\sigma _{T_{V}})\cdot L_T\cdot C_{T_{V}})\nonumber \\{} & {} -CLUP_{T}\cdot C_{T_{V}} +CLUP_T\cdot FR\cdot C_{T_{E,B}} \end{aligned}$$(16) -
3.
Tissue endosome (unbound) \((C_{T_{E,U}})\)
$$\begin{aligned} V_{T_{E}} \cdot \frac{C_{T_{E,U}}}{dt}= & {} CLUP_T\cdot (C_{T_{V}}+C_{T_{I}})-V_{T_{E}}\cdot (kon_{FcRn}\cdot C_{T_{E,U}} \cdot C_{T_{FcRn,U}}\nonumber \\{} & {} -koff_{FcRn}\cdot C_{T_{E,B}}+k_{deg} \cdot C_{T_{E,U}}) \end{aligned}$$(17) -
4.
Tissue endosome (FcRnbound) \((C_{T_{E,B}})\)
$$\begin{aligned} V_{T_{E}} \cdot \frac{C_{T_{E,B}}}{dt}= & {} V_{T_{E}}\cdot (kon_{FcRn}\cdot C_{T_{E,U}} \cdot C_{T_{FcRn,U}}-koff_{FcRn}\cdot C_{T_{E,B}})\nonumber \\{} & {} -CLUP_T\cdot C_{T_{E,B}} \end{aligned}$$(18) -
5.
Tissue interstitium \((C_{T_{I}})\)
$$\begin{aligned} V_{T_{I}} \cdot \frac{C_{T_{I}}}{dt}= & {} (1-\sigma _{T_{V}})\cdot L_T\cdot C_{T_{V}}-(1-\sigma _{T_{L}})\cdot L_T\cdot C_{T_{I}}\nonumber \\{} & {} +CLUP_{T}\cdot (1-FR)\cdot C_{T_{E,B}}-CLUP_T\cdot C_{T_{I}} \end{aligned}$$(19) -
6.
Brain vascular (\(C_{B_{V}}\))
$$\begin{aligned} V_{B_{V}} \cdot \frac{C_{B_{V}}}{dt}= & {} Q_B\cdot C_P-(Q_B-L_B)\cdot C_{B_{V}}-(1-\sigma _{BBB})\cdot Q_{B_{ECF}}\cdot C_{B_{V}} \nonumber \\{} & {} -(1-\sigma _{BCSFB}) \cdot Q_{B_{CSF}}\cdot C_{B_{V}} -CLUP_B\cdot C_{B_{V}} \nonumber \\{} & {} + CLUP_{BBB}\cdot FR_{B} \cdot C_{B_{EBBB,B}} \nonumber \\{} & {} + CLUP_{BCSFB}\cdot FR_{B}\cdot C_{B_{EBCSFB,B}} \end{aligned}$$(20) -
7.
BBB endosome (unbound) (\(C_{B_{EBBB,U}}\))
$$\begin{aligned} V_{B_{EBBB}} \cdot \frac{C_{B_{EBBB,U}}}{dt}= & {} CLUP_{BBB}\cdot (C_{B_{V}}+C_{B_{I}})+V_{B_{EBBB}} \cdot (-kon_{FcRn}\cdot \nonumber \\{} & {} C_{B_{EBBB,U}}\cdot C_{B_{BBBFcRn,U}}+koff_{FcRn}\cdot C_{B_{EBBB,B}} \nonumber \\{} & {} -k_{deg}\cdot C_{B_{EBBB,U}}) \end{aligned}$$(21) -
8.
BBB endosome (FcRnbound) (\(C_{B_{EBB,B}}\))
$$\begin{aligned} V_{B_{EBBB}} \cdot \frac{C_{B_{EBBB,B}}}{dt}= & {} V_{B_{EBBB}} \cdot (kon_{FcRn}\cdot C_{B_{EBBB,U}}\cdot C_{B_{BBBFcRn,U}}\nonumber \\{} & {} -koff_{FcRn}\cdot C_{B_{EBBB,B}})\nonumber \\{} & {} -CLUP_{BBB}\cdot C_{B_{EBBB,B}} \end{aligned}$$(22) -
9.
Brain interstitium (\(C_{B_{{I}}}\))
$$\begin{aligned} V_{B_{I}} \cdot \frac{C_{B_{I}}}{dt}= & {} (1-\sigma _{BBB})\cdot Q_{B_{ECF}}\cdot C_{B_{V}}-(1-\sigma _{B_{ISF}})\cdot Q_{B_{ECF}}\cdot C_{B_{I}}\nonumber \\{} & {} +CLUP_{BBB}\cdot (1-FR_B)\cdot C_{B_{EBBB,B}} -CLUP_{BBB}\cdot C_{B_{I}}\nonumber \\{} & {} -Q_{B_{ECF}}\cdot C_{B_{I}} +Q_{B_{ECF}}\cdot C_{B_{CSF}} \end{aligned}$$(23) -
10.
BCSFB endosome (unbound) (\(C_{B_{EBCSFB,U}}\))
$$\begin{aligned} V_{B_{EBCSFB}}\cdot \frac{C_{B_{EBCSFB,U}}}{dt}= & {} CLUP_{BCSFB}\cdot C_{B_{V}}+CLUP_{BCSFB} \cdot C_{B_{CSF}}\nonumber \\{} & {} +V_{B_{EBCSFB}}\cdot (-kon_{FcRn}\cdot C_{B_{EBCSFB,U}}\nonumber \\{} & {} \cdot C_{B_{BCSFBFcRn,U}}+koff_{FcRn}\cdot C_{B_{EBCSFB,B}}\nonumber \\{} & {} -k_{deg}\cdot C_{B_{EBCSFB,U}}) \end{aligned}$$(24) -
11.
BCSFB endosome (FcRnbound) (\(C_{B_{EBCSFB,B}}\))
$$\begin{aligned} V_{B_{EBCSFB}}\cdot \frac{C_{B_{EBCSFB,B}}}{dt}= & {} V_{B_{EBCSFB}}\cdot (kon_{FcRn}\cdot C_{B_{EBCSFB,U}}\nonumber \\{} & {} \cdot C_{B_{BCSFBFcRn,U}}-koff_{FcRn}\cdot C_{B_{EBCSFB,B}})\nonumber \\{} & {} -CLUP_{BCSFB}\cdot C_{B_{EBCSFB,B}} \end{aligned}$$(25) -
12.
Brain CSF (\(C_{B_{CSF}}\))
$$\begin{aligned} V_{B_{CSF}}\cdot \frac{C_{B_{CSF}}}{dt}= & {} (1-\sigma _{BCSFB})\cdot Q_{B_{CSF}}\cdot C_{B_{V}}-CLUP_{BCSFB} \cdot C_{B_{CSF}}\nonumber \\{} & {} +CLUP_{BCSFB}\cdot (1-FR_B) \cdot C_{B_{EBCSFB,B}} +Q_{B_{ECF}}\cdot C_{B_{I}}\nonumber \\{} & {} -(1-\sigma _{B_{CSF}})\cdot Q_{B_{CSF}} \cdot C_{B_{CSF}}- Q_{B_{ECF}} \cdot C_{B_{CSF}} \end{aligned}$$(26) -
13.
Lymph (\(C_L\))
$$\begin{aligned} V_{L}\cdot \frac{C_{L}}{dt}= & {} (1-\sigma _{T_{L}})\cdot L_T\cdot C_{T_{I}}+(1-\sigma _{B_{CSF}})\cdot Q_{B_{CSF}} \cdot C_{B_{CSF}}\nonumber \\{} & {} + (1-\sigma _{B_{ISF}})\cdot Q_{B_{ECF}}\cdot C_{B_{I}}-(L_T+L_B)\cdot C_L \end{aligned}$$(27) -
14.
FcRn concentration in tissue endosome (\(C_{T_{FcRn,U}}\))
$$\begin{aligned} V_{T_{E}}\cdot \frac{C_{T_{FcRn,U}}}{dt}= & {} -V_{T_{E}}\cdot (kon_{FcRn}\cdot C_{T_{E,U}} \cdot C_{T_{FcRn,U}}-koff_{FcRn}\cdot C_{T_{E,B}})\nonumber \\{} & {} +CLUP_T\cdot C_{T_{E,B}} \end{aligned}$$(28) -
15.
FcRn BBB endosome (\(C_{B_{BBBFcRn,U}}\))
$$\begin{aligned} V_{B_{EBBB}}\cdot \frac{C_{B_{BBBFcRn,U}}}{dt}= & {} V_{B_{EBBB}}\cdot (-kon_{FcRn}\cdot C_{B_{EBBB,U}}\cdot C_{B_{BBBFcRn,U}}\nonumber \\{} & {} +koff_{FcRn}\cdot C_{B_{EBBB,B}})\nonumber \\{} & {} +CLUP_{BBB}\cdot C_{B_{EBBB,B}} \end{aligned}$$(29) -
16.
FcRn BCSFB endosome (\(C_{B_{BCSFBFcRn,U}}\))
$$\begin{aligned} V_{B_{EBCSFB}}\cdot \frac{C_{B_{BCSFBFcRn,U}}}{dt}= & {} V_{B_{EBCSFB}}\cdot (-kon_{FcRn}\cdot C_{B_{EBCSFB,U}}\cdot \nonumber \\{} & {} C_{B_{BCSFBFcRn,U}}+ koff_{FcRn}\cdot C_{B_{EBCSFB,B}}) \nonumber \\{} & {} +CLUP_{BCSFB}\cdot C_{B_{EBCSFB,B}} \end{aligned}$$(30)
This system of equations is augmented with initial conditions. The first concentration is prescribed an initial value \(\frac{Dose_{0}}{V_P}\), where \(Dose_{0}=\frac{Body_{weight} \times Dose_{amount}}{Molecular_{weight}} \times 1000\) with \(Body_{weight}=70 kg\), \(Molecular_{weight}=150kDa\), \(Dose_{amount}=10 mg/kg\). The subsequent 12 concentrations are prescribed initial values of zero, and the remaining three concentrations (\(C_{T_{FcRn,U}}\), \(C_{B_{BBBFcRn,U}}\), \(C_{B_{BCSFBFcRn,U}}\)) are all prescribed a nonzero initial value of \(0.4982\times 10^{-4}.\) In this study, a 10 mg/kg dosage was given over 1000 h to a 70 kg individual using a 1-hour step size.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dadashova, K., Smith, R.C. & Haider, M.A. Local Identifiability Analysis, Parameter Subset Selection and Verification for a Minimal Brain PBPK Model. Bull Math Biol 86, 12 (2024). https://doi.org/10.1007/s11538-023-01234-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-023-01234-4