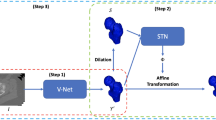
Abstract
Deep Learning (DL) techniques have recently been used in medical image segmentation and the reconstruction of 3D anatomies of a human body. In this work, we propose a semi-supervised DL (SSDL) approach utilizing a CNN-based 3D U-Net model for femur segmentation from sparsely annotated quantitative computed tomography (QCT) slices. Specifically, QCT slices at the proximal end of the femur forming ball and socket joint with acetabulum were annotated for precise segmentation, where a segmenting binary mask was generated using a 3D U-Net model to segment the femur accurately. A total of 5474 QCT slices were considered for training among which 2316 slices were annotated. 3D femurs were further reconstructed from segmented slices employing polynomial spline interpolation. Both qualitative and quantitative performance of segmentation and 3D reconstruction were satisfactory with more than 90% accuracy achieved for all of the standard performance metrics considered. The spatial overlap index and reproducibility validation metric for segmentation—Dice Similarity Coefficient was 91.8% for unseen patients and 99.2% for validated patients. An average relative error of 12.02% and 10.75% for volume and surface area, respectively, were computed for 3D reconstructed femurs. The proposed approach demonstrates its effectiveness in accurately segmenting and reconstructing 3D femur from QCT slices.
Graphical abstract













Similar content being viewed by others
Data availability
The datasets presented during the current study are not publicly available due to privacy and ethical restrictions but might be available on reasonable request from the corresponding author.
References
Sheehan SE, Shyu JY, Weaver MJ, Sodickson AD, Khurana B (2015) Proximal femoral fractures: what the orthopedic surgeon wants to know. Radiographics 35(5):1563–1584
Siebenlist S, Torsiglieri T, Kraus T, Burghardt R, Stöckle U, Lucke M (2010) Comminuted fractures of the proximal ulna—preliminary results with an anatomically preshaped locking compression plate (LCP) system. Injury 41(12):1306–1311
Grignon B, Oldrini G, Walter F (2016) Teaching medical anatomy: what is the role of imaging today? Surg Radiol Anat 38:253–260
Kasban H, El-Bendary M, Salama D (2015) A comparative study of medical imaging techniques. Int J Information Sci Intell Syst 4(2):37–58
Ohnaru K, Sone T, Tanaka K, Akagi K, Ju Y-I, Choi H-J et al (2013) Hip structural analysis: a comparison of DXA with CT in postmenopausal Japanese women. Springerplus 2(1):1–8
Anez-Bustillos L, Derikx LC, Verdonschot N, Calderon N, Zurakowski D, Snyder BD et al (2014) Finite element analysis and CT-based structural rigidity analysis to assess failure load in bones with simulated lytic defects. Bone 58:160–167
Benca E, Synek A, Amini M, Kainberger F, Hirtler L, Windhager R et al (2019) QCT-based finite element prediction of pathologic fractures in proximal femora with metastatic lesions. Sci Rep 9(1):10305
Faisal TR, Luo Y (2017) Study of the variations of fall induced hip fracture risk between right and left femurs using CT-based FEA. Biomed Eng Online 16:1–17
Lee Y, Ogihara N, Lee T (2019) Assessment of finite element models for prediction of osteoporotic fracture. J Mech Behav Biomed Mater 97:312–320
Mirzaei M, Keshavarzian M, Naeini V (2014) Analysis of strength and failure pattern of human proximal femur using quantitative computed tomography (QCT)-based finite element method. Bone 64:108–114
Travascio F, Buller LT, Milne E, Latta L (2021) Mechanical performance and implications on bone healing of different screw configurations for plate fixation of diaphyseal tibia fractures: a computational study. Eur J Orthop Surg Traumatol 31:121–130
Solitro GF, Welborn MC, Mehta AI, Amirouche F (2024) How to optimize pedicle screw parameters for the thoracic spine? A biomechanical and finite element method study. Global. Spine J 14(1):187–194
Solitro GF, Mainnemare F, Amirouche F, Mehta A (2019) A novel technique with reduced computed tomography exposure to predict vertebral compression fracture: a finite element study based on rat vertebrae. Med Biol Eng Comput 57:795–805
Koh K, Kim YH, Kim K, Park WM (2011) Reconstruction of patient-specific femurs using X-ray and sparse CT images. Comput Biol Med 41(7):421–426
Ben Younes L, Nakajima Y, Saito T (2014) Fully automatic segmentation of the femur from 3D-CT images using primitive shape recognition and statistical shape models. Int J Comput Assist Radiol Surg 9:189–196
Carballido-Gamio J, Bonaretti S, Saeed I, Harnish R, Recker R, Burghardt AJ et al (2015) Automatic multi-parametric quantification of the proximal femur with quantitative computed tomography. Quant Imaging Med Surg 5(4):552
Chu C, Bai J, Wu X, Zheng G (2015) MASCG: Multi-atlas segmentation constrained graph method for accurate segmentation of hip CT images. Med Image Anal 26(1):173–184
Xia Y, Fripp J, Chandra SS, Schwarz R, Engstrom C, Crozier S (2013) Automated bone segmentation from large field of view 3D MR images of the hip joint. Phys Med Biol 58(20):7375
Chandra SS, Xia Y, Engstrom C, Crozier S, Schwarz R, Fripp J (2014) Focused shape models for hip joint segmentation in 3D magnetic resonance images. Med Image Anal 18(3):567–578
Xia Y, Chandra SS, Engstrom C, Strudwick MW, Crozier S, Fripp J (2014) Automatic hip cartilage segmentation from 3D MR images using arc-weighted graph searching. Phys Med Biol 59(23):7245
Gilles B, Magnenat-Thalmann N (2010) Musculoskeletal MRI segmentation using multi-resolution simplex meshes with medial representations. Med Image Anal 14(3):291–302
Korfiatis VC, Tassani S, Matsopoulos GK (2017) An independent active contours segmentation framework for bone micro-CT images. Comput Biol Med 87:358–370
Despotović I, Goossens B, Philips W (2015) MRI segmentation of the human brain: challenges, methods, and applications. Comput Math Methods Med 2015
Ahmed SM, Mstafa RJ (2022) A comprehensive survey on bone segmentation techniques in knee osteoarthritis research: from conventional methods to deep learning. Diagnostics. 12(3):611
Besler BA, Michalski AS, Kuczynski MT, Abid A, Forkert ND, Boyd SK (2021) Bone and joint enhancement filtering: application to proximal femur segmentation from uncalibrated computed tomography datasets. Med Image Anal 67:101887
Santarelli C, Argenti F, Uccheddu F, Alparone L, Carfagni M (2020) Volumetric interpolation of tomographic sequences for accurate 3D reconstruction of anatomical parts. Comput Methods Programs Biomed 194:105525
Grassi L, Hraiech N, Schileo E, Ansaloni M, Rochette M, Viceconti M (2011) Evaluation of the generality and accuracy of a new mesh morphing procedure for the human femur. Med Eng Phys 33(1):112–120
Schmid J, Kim J, Magnenat-Thalmann N (2011) Robust statistical shape models for MRI bone segmentation in presence of small field of view. Med Image Anal 15(1):155–168
Kardell M, Magnusson M, Sandborg M, Alm Carlsson G, Jeuthe J, Malusek A (2016) Automatic segmentation of pelvis for brachytherapy of prostate. Radiat Prot Dosimetry 169(1-4):398–404
Vasilache S, Najarian K (2008) Automated bone segmentation from pelvic CT images. In: 2008 IEEE International Conference on Bioinformatics and Biomeidcine Workshops. IEEE, pp 41–47
Kalshetti P, Bundele M, Rahangdale P, Jangra D, Chattopadhyay C, Harit G et al (2017) An interactive medical image segmentation framework using iterative refinement. Comput Biol Med 83:22–33
Stawiaski J, Decenciere E, Bidault F (2008) Interactive liver tumor segmentation using graph-cuts and watershed. In: 11th international conference on medical image computing and computer assisted intervention (MICCAI 2008)
Malusek A, Magnusson M, Sandborg M, Alm CG (2017) A model-based iterative reconstruction algorithm DIRA using patient-specific tissue classification via DECT for improved quantitative CT in dose planning. Med Phys 44(6):2345–2357
Krčah M, Székely G, Blanc R (2011) Fully automatic and fast segmentation of the femur bone from 3D-CT images with no shape prior. In: 2011 IEEE international symposium on biomedical imaging: from nano to macro. IEEE, pp 2087–2090
Lopez-Jimenez F, Attia Z, Arruda-Olson AM, Carter R, Chareonthaitawee P, Jouni H et al (2020) Artificial intelligence in cardiology: present and future. Mayo Clinic Proc 95(5):1015–1039
Peña-Solórzano CA, Albrecht DW, Bassed R, Gillam J, Harris P, Dimmock M (2020) Semi-supervised labelling of the femur in a whole-body post-mortem CT database using deep learning. Comput Biol Med 122:103797
Rokaya D, Kongkiatkamon S, Heboyan A, Dam VV, Amornvit P, Khurshid Z et al (2022) 3d-printed biomaterials in biomedical application. In: Functional Biomaterials: Drug Delivery and Biomedical Applications. Springer, pp 319–339
Luca AR, Ursuleanu TF, Gheorghe L, Grigorovici R, Iancu S, Hlusneac M et al (2022) Impact of quality, type and volume of data used by deep learning models in the analysis of medical images. Inform Med Unlocked 29:100911
Yu AC, Mohajer B, Eng J (2022) External validation of deep learning algorithms for radiologic diagnosis: a systematic review. Radiology. Artificial Intelligence 4(3):e210064
Ronneberger O, Fischer P, Brox T (2015, Proceedings, Part III 18) U-net: convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015. In: 18th International Conference. Springer, Munich, Germany, pp 234–241
Sánchez JCG, Magnusson M, Sandborg M, Tedgren ÅC, Malusek A (2020) Segmentation of bones in medical dual-energy computed tomography volumes using the 3D U-Net. Phys Med 69:241–247
Klein A, Warszawski J, Hillengaß J, Maier-Hein KH (2019) Automatic bone segmentation in whole-body CT images. Int J Comput Assist Radiol Surg 14:21–29
Noguchi S, Nishio M, Yakami M, Nakagomi K, Togashi K (2020) Bone segmentation on whole-body CT using convolutional neural network with novel data augmentation techniques. Comput Biol Med 121:103767
Pham T-T, Le M-B, Le LH, Andersen J, Lou E (2021) Assessment of hip displacement in children with cerebral palsy using machine learning approach. Med Biol Eng Comput 59(9):1877–1887
Wani IM, Arora S (2020) Computer-aided diagnosis systems for osteoporosis detection: a comprehensive survey. Med Biol Eng Comput 58:1873–1917
Chen F, Liu J, Zhao Z, Zhu M, Liao H (2017) Three-dimensional feature-enhanced network for automatic femur segmentation. IEEE J Biomed Health Inform 23(1):243–252
Deng Y, Wang L, Zhao C, Tang S, Cheng X, Deng H-W et al (2022) A deep learning-based approach to automatic proximal femur segmentation in quantitative CT images. Med Biol Eng Comput 60(5):1417–1429
Zhu L, Han J, Guo R, Wu D, Wei Q, Chai W et al (2020) An automatic classification of the early osteonecrosis of femoral head with deep learning. Current medical imaging 16(10):1323–1331
Awal R, Ben Hmida J, Luo Y, Faisal T (2022) Study of the significance of parameters and their interaction on assessing femoral fracture risk by quantitative statistical analysis. Med Biol Eng Comput 60(3):843–854. https://doi.org/10.1007/s11517-022-02516-0
Faisal TR, Luo Y (2016) Study of stress variations in single-stance and sideways fall using image-based finite element analysis. Biomed Mater Eng 27(1):1–14
Wang C, Dahlström N, Fransson S-G, Lundström C (2015) Real-time interactive 3D tumor segmentation using a fast level-set algorithm. J Med Imaging Health Inform 5(8):1998–2002
Khan S, Warkhedkar R, Shyam A (2014) Analysis of Hounsfield unit of human bones for strength evaluation. Procedia Mater Sci 6:512–519
He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In: Proceedings of the IEEE international conference on computer vision, pp 1026–1034
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15(1):1929–1958
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Dubey SR, Singh SK, Chaudhuri BB (2022) Activation functions in deep learning: a comprehensive survey and benchmark. Neurocomputing 503:92–108
Rezatofighi H, Tsoi N, Gwak J, Sadeghian A, Reid I, Savarese S (2019) Generalized intersection over union: a metric and a loss for bounding box regression. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 658–666
Sudre CH, Li W, Vercauteren T, Ourselin S, Jorge CM (2017) Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017. Springer, Québec City, QC, Canada Proceedings 3, pp 240–248
Everingham M, Van Gool L, Williams CK, Winn J, Zisserman A (2010) The pascal visual object classes (voc) challenge. Int J Comput Vis 88:303–338
Ting KM (2010) Sensitivity and specificity. In: Sammut C, Webb GI (eds) Encyclopedia of Machine Learning. Springer US, Boston, MA, pp 901–902
Bjornsson PA, Helgason B, Palsson H, Sigurdsson S, Gudnason V, Ellingsen LM (2021) Automated femur segmentation from computed tomography images using a deep neural network. In: Medical Imaging 2021: Biomedical Applications in Molecular, Structural, and Functional Imaging. SPIE, pp 324–330
Sammut C, Webb GI (2010) Leave-one-out cross-validation. In: Encyclopedia of machine learning, pp 600–601
Kingma DP, Ba J. Adam (2014) A method for stochastic optimization. arXiv preprint arXiv:14126980
Wolberg G, Alfy I (2002) An energy-minimization framework for monotonic cubic spline interpolation. J Comput Appl Math 143(2):145–188
Musy M, Jacquenot G, Dalmasso G, de Bruin R, Pollack A, Claudi F et al (2021) Vedo: a python module for scientific analysis and visualization of 3D objects and point clouds. Zenodo
Chaudhary A, Jhaveri SJ, Sanchez A, Avila LS, Martin KM, Vacanti A et al (2019) Cross-platform ubiquitous volume rendering using programmable shaders in VTK for scientific and medical visualization. IEEE Comput Graph Appl 39(1):26–43
Lorensen WE, Cline HE (1987) Marching cubes: a high resolution 3D surface construction algorithm. ACM Siggraph Computer Graphics 21(4):163–169
Tan Y, Hua J, Dong M (2007) 3D reconstruction from 2D images with hierarchical continuous simplices. Vis Comput 23(9):905–914. https://doi.org/10.1007/s00371-007-0157-0
Guido VR, Drake F Jr (2009) Python 3 reference manual. CreateSpace, Scotts Valley
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C et al (2016) Tensorflow: large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:160304467
Mason D (2011) SU-E-T-33: pydicom: an open source DICOM library. Med Phys 38(6 Part 10):3493
Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D et al (2020) SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17(3):261–272
Zhao C, Keyak JH, Tang J, Kaneko TS, Khosla S, Amin S et al (2020) A deep learning-based method for automatic segmentation of proximal femur from quantitative computed tomography images. arXiv preprint arXiv:200605513
Jeuthe J, Sánchez JCG, Magnusson M, Sandborg M, Tedgren ÅC, Malusek A (2021) Semi-automated 3D segmentation of pelvic region bones in CT volumes for the annotation of machine learning datasets. Radiat Prot Dosimetry 195(3-4):172–176
Acknowledgements
The authors greatly acknowledge Sarah Doll of Mechanical Engineering at UL Lafayette, USA, and Mashiyat Nayeem of Computer Science of North South University, Bangladesh, for helping on data annotation. The authors acknowledge the intellectual contribution of Rabina Awal of Musculoskeletal Mechanics & Multiscale Materials (4M) Lab in the Mechanical Engineering at UL Lafayette throughout the project. The authors also acknowledge the feedback provided by Dr. Ahmed Suparno Bahar Moni, an Assistant Professor and Orthopedic Surgeon in the Dept. of Orthopaedics at the University of Toledo in assessing our annotation.
Author information
Authors and Affiliations
Contributions
TF and MN conceived the idea, and TF, MN, and JS designed the study. JS developed the model and conducted experiments. JS, MN, and TF conducted the data analysis and interpretation of data. All authors discussed the results and contributed to the drafting of this manuscript. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sultana, J., Naznin, M. & Faisal, T.R. SSDL—an automated semi-supervised deep learning approach for patient-specific 3D reconstruction of proximal femur from QCT images. Med Biol Eng Comput 62, 1409–1425 (2024). https://doi.org/10.1007/s11517-023-03013-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-03013-8