Abstract
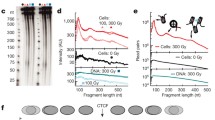
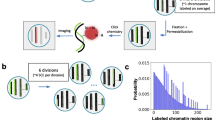
Spatial chromatin structure plays fundamental roles in many vital biological processes including DNA replication, transcription, damage and repair. However, the current understanding of the secondary structure of chromatin formed by local nucleosome-nucleosome interactions remains controversial, especially for the existence and conformation of 30 nm structure. Since chromatin structure influences the fragment length distribution (FLD) of ionizing radiation-induced DNA strand breaks, a 3D chromatin model fitting FLD patterns can help to distinguish different models of chromatin structure. Here, we developed a novel “30-C” model combining 30 nm chromatin structure models with Hi-C data, which measured the spatial contact frequency between different loci in the genome. We first reconstructed the 3D coordinates of the 25 kb bins from Hi-C heatmaps. Within the 25 kb bins, lower level chromatin structures supported by recent studies were filled. Simulated FLD patterns based on the 30-C model were compared to published FLD patterns induced by heavy ion radiation to validate the models. Importantly, the 30-C model predicted that the most probable chromatin fiber structure for human interphase fibroblasts in vivo was 45% zig-zag 30 nm fibers and 55% 10 nm fibers.
Similar content being viewed by others

References
Abbas, A., He, X., Niu, J., Zhou, B., Zhu, G., Ma, T., Song, J., Gao, J., Zhang, M.Q., and Zeng, J. (2019). Integrating Hi-C and FISH data for modeling of the 3D organization of chromosomes. Nat Commun 10, 2049.
Albiez, H., Cremer, M., Tiberi, C., Vecchio, L., Schermelleh, L., Dittrich, S., Küpper, K., Joffe, B., Thormeyer, T., von Hase, J., et al. (2006). Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res 14, 707–733.
Botchway, S.W., Stevens, D.L., Hill, M.A., Jenner, T.J., and O’Neill, P. (1997). Induction and rejoining of DNA double-strand breaks in Chinese hamster V79-4 cells irradiated with characteristic aluminum K and copper L ultrasoft X rays. Radiat Res 148, 317–324.
Boudaïffa, B., Cloutier, P., Hunting, D., Huels, M.A., and Sanche, L. (2000). Resonant formation of DNA strand breaks by low-energy (3 to 20 eV) electrons. Science 287, 1658–1660.
Bryant, P.E. (1985). Enzymatic restriction of mammalian cell DNA: Evidence for double-strand breaks as potentially lethal lesions. Int J Radiat Biol Related Studies Phys Chem Med 48, 55–60.
Cremer, T., and Cremer, C. (2001). Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2, 292–301.
de Lara, C.M., Hill, M.A., Jenner, T.J., Papworth, D., and O’Neill, P. (2001). Dependence of the yield of DNA double-strand breaks in Chinese hamster V79-4 cells on the photon energy of ultrasoft X rays. Radiat Res 155, 440–448.
Dekker, J., Rippe, K., Dekker, M., and Kleckner, N. (2002). Capturing chromosome conformation. Science 295, 1306–1311.
Dixon, J.R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., Hu, M., Liu, J. S., and Ren, B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380.
Dostie, J., Richmond, T.A., Arnaout, R.A., Selzer, R.R., Lee, W.L., Honan, T.A., Rubio, E.D., Krumm, A., Lamb, J., Nusbaum, C., et al. (2006). Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res 16, 1299–1309.
Du, Z., Zheng, H., Huang, B., Ma, R., Wu, J., Zhang, X., He, J., Xiang, Y., Wang, Q., Li, Y., et al. (2017). Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547, 232–235.
Dutuit, O., Tabche-Fouhaile, A., Nenner, I., Frohlich, H., and Guyon, P.M. (1985). Photodissociation processes of water vapor below and above the ionization potential. J Chem Phys 83, 584–596.
Eltsov, M., Maclellan, K.M., Maeshima, K., Frangakis, A.S., and Dubochet, J. (2008). Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA 105, 19732–19737.
Finch, J.T., and Klug, A. (1976). Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA 73, 1897–1901.
Frankenberg, D., Frankenberg-Schwager, M., Blöcher, D., Harbich, R., and Blocher, D. (1981). Evidence for DNA double-strand breaks as the critical lesions in yeast cells irradiated with sparsely or densely ionizing radiation under oxic or anoxic conditions. Radiat Res 88, 524–532.
Friedland, W., Dingfelder, M., Kundrát, P., and Jacob, P. (2011). Track structures, DNA targets and radiation effects in the biophysical Monte Carlo simulation code PARTRAC. Mutat Res 711, 28–40.
Friedland, W., Jacob, P., Paretzke, H.G., Merzagora, M., and Ottolenghi, A. (1999). Simulation of DNA fragment distributions after irradiation with photons. Radiat Environ Biophys 38, 39–47.
Friedland, W., and Kundrát, P. (2013). Track structure based modelling of chromosome aberrations after photon and alpha-particle irradiation. Mutat Res 756, 213–223.
Friedland, W., Paretzke, H.G., Ballarini, F., Ottolenghi, A., Kreth, G., and Cremer, C. (2008). First steps towards systems radiation biology studies concerned with DNA and chromosome structure within living cells. Radiat Environ Biophys 47, 49–61.
Fussner, E., Ching, R.W., and Bazett-Jones, D.P. (2011). Living without 30 nm chromatin fibers. Trends Biochem Sci 36, 1–6.
Ghirlando, R., and Felsenfeld, G. (2008). Hydrodynamic studies on defined heterochromatin fragments support a 30-nm fiber having six nucleosomes per turn. J Mol Biol 376, 1417–1425.
Grigoryev, S.A., Arya, G., Correll, S., Woodcock, C.L., and Schlick, T. (2009). Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc Natl Acad Sci USA 106, 13317–13322.
Hall, E.J., and Giaccia, A.J. (2011). Radiobiology for the Radiologist, 7th ed. (Lippincott Williams & Wilkins).
Höglund, E., Blomquist, E., Carlsson, J., and Stenerlöw, B. (2000). DNA damage induced by radiation of different linear energy transfer: initial fragmentation. Int J Radiat Biol 76, 539–547.
Höglund, E., and Stenerlöw, B. (2001). Induction and rejoining of DNA double-strand breaks in normal human skin fibroblasts after exposure to radiation of different linear energy transfer: Possible roles of track structure and chromatin organization. Radiat Res 155, 818–825.
Hsieh, T.H.S., Weiner, A., Lajoie, B., Dekker, J., Friedman, N., and Rando, O.J. (2015). Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell 162, 108–119.
Imakaev, M., Fudenberg, G., McCord, R.P., Naumova, N., Goloborodko, A., Lajoie, B.R., Dekker, J., and Mirny, L.A. (2012). Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods 9, 999–1003.
Incerti, S., Baldacchino, G., Bernal, M., Capra, R., Champion, C., Francis, Z., Guèye, P., Mantero, A., Mascialino, B., Moretto, P., et al. (2010a). The Geant4-DNA project. Int J Model Simul Sci Comput 01, 157–178.
Incerti, S., Ivanchenko, A., Karamitros, M., Mantero, A., Moretto, P., Tran, H.N., Mascialino, B., Champion, C., Ivanchenko, V.N., Bernal, M.A., et al. (2010b). Comparison of GEANT4 very low energy cross section models with experimental data in water. Med Phys 37, 4692–4708.
Li, G., Fullwood, M.J., Xu, H., Mulawadi, F.H., Velkov, S., Vega, V., Ariyaratne, P.N., Mohamed, Y.B., Ooi, H.S., Tennakoon, C., et al. (2010). ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol 11, R22.
Liang, Y., Fu, Q., Wang, X., Liu, F., Yang, G., Luo, C., Ouyang, Q., and Wang, Y. (2017). Relative biological effectiveness for photons: implication of complex DNA double-strand breaks as critical lesions. Phys Med Biol 62, 2153–2175.
Liang, Y., Yang, G., Liu, F., and Wang, Y. (2016). Monte Carlo simulation of ionizing radiation induced DNA strand breaks utilizing coarse grained high-order chromatin structures. Phys Med Biol 61, 445–460.
Lieberman-Aiden, E., van Berkum, N.L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., Amit, I., Lajoie, B.R., Sabo, P.J., Dorschner, M.O., et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293.
Löbrich, M., Cooper, P.K., and Rydberg, B. (1996). Non-random distribution of DNA double-strand breaks induced by particle irradiation. Int J Radiat Biol 70, 493–503.
Luger, K., Dechassa, M.L., and Tremethick, D.J. (2012). New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 13, 436–447.
Luger, K., Mäder, A.W., Richmond, R.K., Sargent, D.F., and Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260.
Maeshima, K., Hihara, S., and Eltsov, M. (2010). Chromatin structure: does the 30-nm fibre exist in vivo? Curr Opin Cell Biol 22, 291–297.
Martin, F., Burrow, P.D., Cai, Z., Cloutier, P., Hunting, D., and Sanche, L. (2004). DNA strand breaks induced by 0–4 eV electrons: the role of shape resonances. Phys Rev Lett 93, 068101.
Newman, H.C., Prise, K.M., Folkard, M., and Michael, B.D. (1997). DNA double-strand break distributions in X-ray and alpha-particle irradiated V79 cells: Evidence for non-random breakage. Int J Radiat Biol 71, 347–363.
Nikjoo, H., O’Neill, P., Terrissol, M., and Goodhead, D.T. (1999). Quantitative modelling of DNA damage using Monte Carlo track structure method. Radiat Environ Biophys 38, 31–38.
Nygren, J., Ljungman, M., and Ahnström, G. (1995). Chromatin structure and radiation-induced DNA strand breaks in human cells: soluble scavengers and DNA-bound proteins offer a better protection against single- than double-strand breaks. Int J Radiat Biol 68, 11–18.
Ohno, M., Ando, T., Priest, D.G., Kumar, V., Yoshida, Y., and Taniguchi, Y. (2019). Sub-nucleosomal genome structure reveals distinct nucleosome folding motifs. Cell 176, 520–534.e25.
Ohno, M., Priest, D.G., and Taniguchi, Y. (2018). Nucleosome-level 3D organization of the genome. Biochem Soc Trans 46, 491–501.
Oleinick, N.L., Chiu, S.M., Ramakrishnan, N., and Xue, L.Y. (1987). The formation, identification, and significance of DNA-protein cross-links in mammalian cells. Br J Cancer Suppl 8, 135–140.
Oluwadare, O., Highsmith, M., and Cheng, J. (2019). An overview of methods for reconstructing 3-D chromosome and genome structures from Hi-C data. Biol Proced Online 21, 7.
Ou, H.D., Phan, S., Deerinck, T.J., Thor, A., Ellisman, M.H., and O’Shea, C.C. (2017). ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, eaag0025.
Pal, K., Forcato, M., and Ferrari, F. (2019). Hi-C analysis: from data generation to integration. Biophys Rev 11, 67–78.
Prise, K.M., Folkard, M., Davies, S., and Michael, B.D. (1989). Measurement of DNA damage and cell killing in Chinese hamster V79 cells irradiated with aluminum characteristic ultrasoft X rays. Radiat Res 117, 489–499.
Rao, S.S.P., Huntley, M.H., Durand, N.C., Stamenova, E.K., Bochkov, I.D., Robinson, J.T., Sanborn, A.L., Machol, I., Omer, A.D., Lander, E.S., et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680.
Ricci, M.A., Manzo, C., García-Parajo, M.F., Lakadamyali, M., and Cosma, M.P. (2015). Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160, 1145–1158.
Risca, V.I., Denny, S.K., Straight, A.F., and Greenleaf, W.J. (2017). Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping. Nature 541, 237–241.
Roots, R., and Okada, S. (1975). Estimation of life times and diffusion distances of radicals involved in X-ray-induced DNA strand breaks or killing of mammalian cells. Radiat Res 64, 306–320.
Rydberg, B. (1996). Clusters of DNA damage induced by ionizing radiation: Formation of short DNA fragments. II. Experimental detection. Radiat Res 145, 200–209.
Rydberg, B., Holley, W.R., Mian, I.S., and Chatterjee, A. (1998). Chromatin conformation in living cells: support for a zig-zag model of the 30 nm chromatin fiber. J Mol Biol 284, 71–84.
Simonis, M., Klous, P., Splinter, E., Moshkin, Y., Willemsen, R., de Wit, E., van Steensel, B., and de Laat, W. (2006). Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38, 1348–1354.
Song, F., Chen, P., Sun, D., Wang, M., Dong, L., Liang, D., Xu, R.M., Zhu, P., and Li, G. (2014). Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–380.
Stevens, T.J., Lando, D., Basu, S., Atkinson, L.P., Cao, Y., Lee, S.F., Leeb, M., Wohlfahrt, K.J., Boucher, W., O’Shaughnessy-Kirwan, A., et al. (2017). 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64.
Wang, S., Su, J.H., Beliveau, B.J., Bintu, B., Moffitt, J.R., Wu, C., and Zhuang, X. (2016). Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602.
Wu, P., Li, T., Li, R., Jia, L., Zhu, P., Liu, Y., Chen, Q., Tang, D., Yu, Y., and Li, C. (2017). 3D genome of multiple myeloma reveals spatial genome disorganization associated with copy number variations. Nat Commun 8, 1937.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) (11875079 and 11434001). C.L. was supported by NSFC (31871266), the National Key Research and Development Project of China (2016YFA0100103), and NSFC Key Research Grant 71532001.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Compliance and ethics The author(s) declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Ma, W., Gu, C., Ma, L. et al. Mixed secondary chromatin structure revealed by modeling radiation-induced DNA fragment length distribution. Sci. China Life Sci. 63, 825–834 (2020). https://doi.org/10.1007/s11427-019-1638-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-019-1638-6