Abstract
The Anatolian peninsula otherwise known as Asia Minor is considered one of the centers that shaped grape (Vitis spp.) evolution and domestication. This region with diverse ecological conditions also has a long history of viticulture and growing grapes has been a part of the local culture since very old times. However, very little information is available on genetic analysis of Anatolian grape germplasm. This study reports on genetic analyses of 88 grapevine cultivars from Central Anatolia using 17 microsatellite (SSR) loci. The average number of alleles per locus was 9.18, ranging from 5 to 15. The highest heterozygosity rate was obtained for the SSR loci “VVS2” and “VMC2H4.” Genetic distances between populations ranged from 0.056 to 0.207 and two cases of identical, seven cases of homonymous, and nine cases of synonymous grape cultivar groups were identified. Based on comparisons with international Vitis databases, it has been determined that “Moldova-Coarna Neagra” cultivar is synonymous with some Anatolian cultivars. In addition, investigation of the genetic diversity of 20 genotypes of Anatolian wild germplasm revealed a higher level of genetic diversity in wild populations than in cultivated ones at the studied microsatellite loci. The results reported here should not only contribute towards better management of the grape germplasm of the region but also provide new insights into grape domestication.



Similar content being viewed by others
References
Almadanim MC, Baleiras-Couto MM, Pereira HS, Carneiro LC, Fevereiro P, Eiras-Dias JE, Morais-Cecilio L, ViegasmW VMM (2007) Genetic diversity of the grapevine (Vitis vinifera L.) cultivars most utilized for wine production in Portugal. Vitis 46(3):116–119
Arroyo-García R, Revilla E (2013) The current status of wild grapevine populations (Vitis vinifera ssp sylvestris) in the Mediterranean basin. https://doi.org/10.5772/52933
Arroyo-Garcia R, Ruiz-Garcia L, Boulling L, Ocete R, López MA, Arnold C, Ergul A, Söylemezoğlu G, Uzun Hİ, Cabello F, Ibáñez J, Aradhya MK, Atanassov A, Atanassov I, Balint S, Cenis JL, Costantini L, Gorislavets S, Grando MS, Klein BY, McGovern P, Merdinoglu D, Pejic I, Pelsy F, Primikirios N, Risovannaya V, Roubelakis-Angelakis KA, Snouss H, Sotiri P, Tamhankar S, This P, Troshin L, Malpica JM, Lefort F, Martinez-Zapater JM (2006) Multiple origins of cultivated grapevine (Vitis vinifera L. ssp.sativa) based on chloroplast DNA polymorphisms. Mol Ecol 15(12):3707–3714
Barnaud A, Laucou V, This P, Lacombe T, Doligez A (2010) Linkage disequilibrium in wild French grapevine, Vitis vinifera L subsp silvestris. Heredity 104:431–437
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457
Bowers JE, Dangl GS, Meredith CP (1999) Development and characterization of additional microsatellite DNA markers for grape. Am J Enol Vitic 50(3):243–246
Boz Y, Bakır M, Çelikkol BP, Kazan K, Yılmaz F, Çakır B, Aslantaş Ş, Söylemezoğlu G, Yaşasın AS, Özer C, Çelik H, Ergül A (2011) Genetic characterization of grape (Vitis vinifera L.) germplasm from Southeast Anatolia by SSR markers. Vitis 50(3):99–106
Corander J, Marttinen P, Siren J, Tang J, Cheng L (2005-2012) Bayesian analysis of population structure (BAPS), version 6.0. http://www.Helsinki.fi/bsg/software/BAPS
Corander J, Marttinen P, Sirén J, Tang J (2008) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9:539
Crespan M, Cancellier S, Costacurta A, Guist M, Carraro R, Stefano R, Santangelo S (2003) Contribution to the clearing up of synonymies in some groups of Italian grapevine cultivars. Proc Acta Hortic 603:275–289
Cunha J, Balerias-Couto M, Cunha JP, Banza J, Soveral A, Carneiro LC, Eiras-Dias JE (2007) Characterization of Portuguese populations of Vitis vinifera L ssp sylvestris (Gmelin) Hegi. Genet Resour Crop Evol 54:981–988. https://doi.org/10.1007/s10722-006-9189-y
Cutanda-Perez MC, Ageorges A, Gomez C, Vialet S, Terrier N, Romieu C, Torregrosa L (2009) Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol Biol 69:633–648
de Andrés MT, Benito A, Pérez-Rivera G, Ocete R, Lopez MA, Gaforio L, Muñoz G, Cabello F, Martinez-Zapater JM, Arroyo-Garcia R (2012) Genetic diversity of wild grapevine populations in Spain and their genetic relationships with cultivated grapevines. Mol Ecol 21:800–816
Dettweiller E, Jung A, Zyprian E, Töpfer R (2000) Grapevine cultivar Müller-Thurgau and its true to type descent. Vitis 39(2):63–65
Di Vecchi-Staraz M, Lucou V, Bruno G, Lamcombe T, Gerber S, Bourse T, Boselli M, This P (2009) Low level of pollen-mediated gene flow from cultivated to wild grapevine: consequences for the evolution of the endangered subspecies Vitis vinifera L. ssp silvestris. J Hered 100(1):66–75. https://doi.org/10.1093/jhered/esn084
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martinez-Zapater JM, Zyprian E, Moreira FM (2013) Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol 13(39):1–17
Ergül A, Kazan K, Aras S, Çevik V, Çelik H, Söylemezoğlu G (2006) AFLP analysis of genetic variation within the two economically important Anatolian grapevine (Vitis viniferaL.) varietal groups. Genome 49:467–495
Ergül A, Perez-Rivera G, Söylemezoğlu G, Kazan K, Arroyo-Garcia R (2011) Genetic diversity in Anatolian wild grapes (Vitis vinifera subsp. sylvestris) estimated by SSR markers. Plant Genet Resour 9(3):375–383
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resour 10:564–567
Eyduran PS, Ercisli S, Akin M, Eyduran E (2015) Genetic characterization of autochthonous grapevine cultivars from Eastern Turkey by simple sequence repeats (SSRs) ARTICLE; Agriculture and Environmental Biotechnology. https://doi.org/10.1080/13102818.2015.1105726
Ferrara G, Gallotta A, Pacucci C, Matarrese AMS, Mazzeo A, Giancaspro A, Gadaleta A, Piazzolla F, Colelli G (2017) The table grape ‘Victoria’ with a long shaped berry: a potential mutation with attractive characteristics for consumers. J Sci Food Agric. https://doi.org/10.1002/jsfa.8429
Gismondi A, Impei S, Di Marco G, Crespan M, Leonardi D, Canini A (2014) Detection of new genetic profiles and allelic variants in improperly classified grapevine accessions. Genome 57:111–118
Goto-Yamamoto N, Mouri H, Azumi M, Edwards KJ (2006) Development of grape microsatellite markers and mikrosatellite analysis including oriental cultivars. Am J Enol Vitic 57(1):105–108
Goto-Yamamoto N, Azuma A, Mitani N, Kobayash S (2013) SSR genotyping of wild grape species and grape cultivars of Vitis vinifera and V. vinifera×V. labrusca. J Japan Soc Hort Sci 82(2):125–130
Grassi F, Labra M, Imazio S, Spada A, Sgorbati S, Scienza A, Sala F (2003) Evidence of a secondary grapevine domestication center detected by SSR analysis. Theor Appl Genet 107:1315–1320
Grassi F, De Mattia F, Zecca G, Sala F, Labra M (2008) Historical isolation and quaternary range expansion of divergent lineages in wild grapevine. Biol J Linn Soc 95:611–619
Guo DL, Yu YH, Xi FF, Shi YY, Zhang GH (2016) Histological and molecular characterization of grape early ripening bud mutant. Int J Genomıcs 2016:1–7
Hizarci Y, Ercisli S, Yuksel C, Ergul A (2012) Genetic characterization and relatedness among autochthonous grapevine cultivars from Northeast Turkey by simple sequence repeats (SSR). J Appl Bot Food Qual 85:224–228
Hvarleva T, Rusanov K, Lefort F, Tsvetkov I, Atanassov A, Atanassov I (2004) Genotyping of Bulgarian Vitis vinifera L. cultivars by microsatellite analysis. Vitis 43:27–34
Jin W, Wang H, Li M, Wang J, Yang Y, Zhang X, Yan G, Zhang H, Liu J, Zhang K (2016) The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunusavium L.). Plant Biotechnol J 14(11):2120–2133
Karataş H, Değirmenci D, Velasco R, Vezzulli S, Bodur Ç, Ağaoğlu YS (2007) Microsatellite fingerprinting of homonymous grapevine (Vitis vinifera L.) varieties in neighboring regions of south-East Turkey. Sci Hortic. https://doi.org/10.1016/j.scienta.2007.07.001
Karataş DD, Karataş H, Laucou V, Sarikamiş G, Riahi L, Bacilieri R, This P (2014) Genetic diversity of wild and cultivated grapevine accessions from Southeast Turkey. Hereditas 151:73–80
Khadivi A, Gismondi A, Canini A (2017) Genetic characterization of Iranian grapes (Vitis vinifera L.) and their relationships with Italian ecotypes. Agrofor Syst. https://doi.org/10.1007/s10457-017-0134-1
Labra M, Imazio S, GrassI F, RossonI M, Sala F (2004) Vine-1 retrotransposon-based sequence-specific amplified polymorphism for Vitis vinifera L. genotyping. Plant Breed 123:180–185
Lacombe T, Boursiquot JM, Laucou V, Di Vecchi-Staraz M, Péros JP, This P (2013) Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor Appl Genet 126:401–414
Laucou V, Lacombe T, Dechesne F, Siret R, Bruno JP, Dessup M, Dessup T, Ortigosa P, Parra P, Roux C, Santoni S, Varès D, Péros JP, Boursiquot JM, This P (2011) High throughput analysis of grape genetic diversity as a tool for germplasm collection management. Theor Appl Genet 122:1233–1245
Lefort F, Lally M, Thompson D, Douglas GC (1998) Morphologicaltraits, microsatellite fingerprinting and genetic relatedness of a stand of elite oaks (Q. robur L.) at Tullynally, Ireland. Silvae Genetica 47:257–262
Lopes MS, Mendonça D, Rodrigues dos Santos M, Eiras-Dias JE, da Câmara MA (2009) New insights on the genetic basis of Portuguese grapevine and on grapevine domestication. Genome 52:790–800
Maul E, Sudharma KN, Kecke S, Marx G, Müller C, Audeguin L, … This P (2012) The European Vitis database (www.eu-vitis.de) - a technical innovation through an online uploading and interactive modification system. Vitis 51:79–86
Maul E, Eibach R, Zyprian E, Töpfer R (2015) The prolific grape variety (Vitis vinifera L.) ‘Heunisch Weiss’ (= ‘Gouaisblanc’): bud mutants, “colored” homonyms and further offspring. Vitis 54:79–86
Mcgovern P (2003) Ancient wine, the search for the origins of viticulture. Princeton University Press, Princeton
Medhi K, Sarmah DK, Deka M, Bhau BS (2014) High gene flow and genetic diversity in three economically important Zanthoxylum Spp. of Upper Brahmaputra Valley Zone of NE India using molecular markers. Meta Gene:706–721
Meneghetti S, Costacurta A, Frare E, Da Rold G, Migliaro D, Morreale G, Crespan M, Sotés V, Calò A (2011) Clones identification and genetic characterization of Garnacha grapevine by means of different PCR-derived marker systems. Mol Biotechnol 48(3):244–254
Merdinoglu D, Butterlin G, Baur C, Balthazard J, Bouquet A, Boursiquot JM (2000) Comparison of RAPD, AFLP and SSR (microsatellite) markers for genetic diversity analysis in Vitis vinifera L. Acta Hortic 528:193–197
Minch E, Ruiz-Linares A, Goldstein DB, Feldman M, Cavalli-Sforza LL (1995) Microsat (version 1.4d): a computer program for calculating various statistics on microsatellite allele data. http://lotka.stanford.edu:microsat.html.
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Olmo HP (1935) Empty seededness in varieties of Vitis vinifera. Proceedings of the J Am Soc Hortıc Scı 32:376–380
Regner F, Hack R, Santiago JL (2006) Highly variable Vitis microsatellite loci for the identification of pinot noir clones. Vitis 45(2):85–91
Riahi L, Soghlami N, El-Heir K, Laucou V, Cunff LL, Boursiquot JM, Lacombe T, Mliki A, Ghorbel A, This P (2010) Genetic structure and differentiation among grapevines (Vitis vinifera) accessions from Maghred region. Genet Resour Crop Evol 57:255–272
Riaz S, De Lorenzis G, Velasco D, Koehmstedt A, Maghradze D, Bobokashvili Z, Musayev M, Zdunic G, Laucou V, Walker MA, Failla O, Preece JE, Aradhya M, Arroyo-Garcia R (2018) Genetic diversity analysis of cultivated and wild grapevine (Vitis vinifera L.) cultivars around the Mediterranean basin and Central Asia. BMC Plant Biol 18(137):1–14. https://doi.org/10.1186/s12870-018-1351-0
Rohlf F (1988) NTSYS-PC numerical taxonomy and multivariate analysis system, version 2.0. Setoukat, NY, USA
Rohlf F (2004) NTSYS-pc: numeric taxonomic systems. Applied biostatistics Inc. version 2.20. Setoukat, NY, USA
Salayeva S, Decroocq S, Mariette S, Akhundova E (2010) Comparıison of genetic diversity between cultivated and wild grape varieties originating from the near-Caspian zone of Azerbaijan. J Int Sci Vigne Vin 44(4):191–200
Sefc KM, Lopes MS, Lefort F, Botta R, Roubelakis-Angelakis KA, Ibañez J, Pejic I, Wegner HW, Glössl J, Steinkellner H (2000) Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theor Appl Genet 100:498–505
Selli F, Bakir M, Inan G, Aygun H, Boz Y, Yasasin AS, Ozer X, Akman B, Soylemezlu G, Kazan K, Ergul A (2007) Simple sequence repeat based assessment of genetic diversity in Dimrit and Germe grapevine cultivars from Turkey. Vitis 46:182–187
Slatkin M, Barton NH (1989) A comparison of three indirect methods for estimating the average level of gene flow. Evolution 43:1349–1368
Stajner N, Tomić L, Ivanisević D, Korać N, Cvetković-Jovanović T, Beleski K, Angleova E, Maraš V, Javornik B (2014) Microsatellite inferred genetic diversity and structure of Western Balkan grapevines (Vitis vinifera L.). Tree Genet Genomes 10:127–140
Tangolar SG, Soydam S, Bakir M, Karaaǧaç E (2009) Genetic analysis of grapevine cultivars from the eastern Mediterranean region of Turkey, based on SSR markers. J Agric Sci 15:1–8
Terral JF, Tabard E, Bouby L, Ivorra S, Pastor T, Figueiral I, Picq S, Chevance JB, Jung C, Fabre L, Tardy C (2010) Evolution and history of grapevine (Vitis vinifera) under domestication: new morphometric perspectives to understand seed domestication syndrome and reveal origins of ancient European cultivars. Ann Bot 105:443–455
This P, Lacombe T, Thomas MR (2006) Historical origins and genetic diversity of wine grapes. Trends Genet 22:511–519
Thomas MR, Scott NS (1993) Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequenced-tagged sites (STSs). Theor Appl Genet 86:985–990
Veron V, Caron H, Degen B (2005) Gene flow and mating system of the tropical tree Sextonia rubra. Silvae Genetica 54:275–280
Villano C, Carputo D, Frusciante L, Santoro X, Aversano R (2014) Use of SSR and Retrotransposon-based markers to interpret the population structure of native grapevines from southern Italy. Mol Biotechnol 56:1011–1020. https://doi.org/10.1007/s12033-014-9780-y
Vouillamoz JF, McGovern PE, Ergul A, Söylemezoğlu G, Tevzadze G, Meredith CP, Grando MS (2006) Genetic characterization and relationships of traditional grape cultivars from Transcaucasia and Anatolia. Plant Genet Resour 4(2):144–158
Wagner HW, Sefc KM (1999) IDENTITY 1.0. Centre for Applied Genetics. University of Agricultural Science, Vienna
Walker AR, Lee E, Bogs J, McDavid DAJ, Thomas MR, Robinson SP (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49:772–785
Wang Z, Kang M, Liu H, Gao J, Zhang Z, Li Y, Wu R, Pang X (2014) High-level genetic diversity and complex population structure of Siberian apricot (Prunus sibirica L.) in China as revealed by nuclear SSR markers. PLoS One 9(2):e87381. https://doi.org/10.1371/journal.pone.0087381
Wang L, Zhang J, Liu L, Zhang L, Wei L, Hu D (2015) Genetic diversity of grape germplasm as revealed by microsatellite (SSR) markers. Afr J Biotechnol 14(2):990–998
Yang H, Zhang R, Jin G, Feng Z, Zhou Z (2016) Assessing the genetic diversity and genealogical reconstruction of cypress (CupressusfunebrisEndl.) breeding parents using SSR markers. Forests 7(160):1–14. https://doi.org/10.3390/f7080160
Zinelabidine LH, Haddioui A, Bravo G, Arroyo-Garcia R, Martinez-Zapater JM (2010) Genetic origins of cultivated and wild grapevines from Morocco. Am J Enol Vitic 61:83–90
Funding
This study was supported by the Scientific and Technical Research Council of Turkey (TUBITAK) (Grant No. 105 G 078).
Author information
Authors and Affiliations
Contributions
AE designed and outlined the research. FY, MS, CYÖ, and GS conducted the experiments. KK, NH, FY, and CYÖ performed the bioinformatics studies, interpreted the data, and wrote the manuscript. TU, CÖ, ASY, YB, and HÇ organized the plant collection and preparation. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
The names of internationally known SSR loci as well as ampelographic and molecular data from the grape cultivars used in the study were presented in the text and in the Supplementary Tables.
Additional information
Communicated by G. G. Vendramin
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Table 1
Names, cultivar numbers, bunch-berry properties and ripening times of the grapevine cultivars from Central Anatolia (DOCX 46 kb).
Supplementary Table 2
Allele sizes (bp) of grape Central Anatolian and reference cultivars at 17 SSR loci (See Supplementary Table 1 for corresponding cultivar no) (DOCX 94 kb).
Supplementary Table 3
Central Anatolian grape cultivars with varying degree of genetic similarity identified based on SSR analysis. Cultivar name (Cultivar no, See Supplementary Table 1 for corresponding cultivar no) (DOCX 30 kb).
Supplementary Table 4
Expected (He) and observed (Ho) heterozygosity values for Central Anatolian cultivars, wild grapes and reference group (DOCX 14 kb).
Supplementary Table 5
Genetic differentiation (Fst, below diagonal) and gene flow (Nm, above diagonal) between Central Anatolian cultivars, wild grapes and reference group (DOCX 12 kb).
Supplementary Table 6
Nei’s genetic distance values measured between cultivars, wild grapes and reference group (DOCX 15 kb).
Supplementary Figure 1
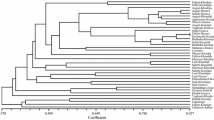
Genetic relationships observed between Central Anatolian cultivars-wild grape populations. The dendrogram was constructed by neighbor-joining clustering based on Nei’s (1972) genetic distance (JPG 26 kb).
Supplementary Figure 2
Bayesian analysis of population structure (BAPS) of Central Anatolian cultivars. A. Grape cultivars from Central Anatolia were grouped according to provinces and subjected to BAPS analysis to observe population structuring. B. The same analysis was repeated based on the individual cultivars. Each vertical bar represents one individual genotype, populations are separated by black vertical lines (JPG 73 kb).
Rights and permissions
About this article
Cite this article
Yılmaz, F., Shidfar, M., Hazrati, N. et al. Genetic analysis of central Anatolian grapevine (Vitis vinifera L.) germplasm by simple sequence repeats. Tree Genetics & Genomes 16, 55 (2020). https://doi.org/10.1007/s11295-020-01429-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-020-01429-z