Abstract
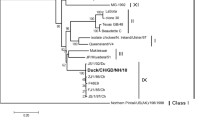
Newcastle disease (ND) is one of the most serious diseases affecting poultry worldwide. In 2022, we studied two strains of Newcastle disease virus (NDV) from pigeons and magpies identified by PCR and propagated in SPF chicken embryos. The whole genome of the virus was then expanded and its biological characteristics were studied. The results showed that NDV was isolated from pigeons and magpies. Virus present in the allantoic fluid could agglutinate red blood cells and could not be neutralized by serum positive for avian influenza. Sequencing showed that the gene length of the two isolates was 15,191 bp, had high homology and was located in the same branch of the phylogenetic tree, both belonging to genotype VI.1.1. The sequence of 112–117 amino acids in the F gene sequence was 112R-R-Q-K-R-F117, which constituted virulent strain characteristics. The HN gene contained 577 amino acids, which is also consistent with the characteristics of a virulent strain. The results from the study of biological characteristics revealed that the virulence of SX/TY/Pi01/22 was slightly stronger. There were only four different bases in the complete sequence of the two strains. Comprehensive analysis revealed that the G at 11,847 site of the SX/TY/Ma01/22 strain may change to T, leading to translation of amino acids from R to S, thereby weakening viral virulence. Therefore, NDV was transmitted from pigeons to magpies, indicating that the pathogen could be transmitted between poultry and wild birds.





Similar content being viewed by others
References
World Organization for Animal Health (2022) Newcastle disease. WOAH. https://www.woah.org/en/disease/newcastle-disease/. Accessed 26 December 2022
Notice of the Ministry of Agriculture and Rural affairs of People’s Republic of China (2022) Lists A, B and C Diseases. Ministry of Agriculture and Rural affairs of the People’s Republic of China. https://www.springer.com/journal/11262/submission-guidelines. Accessed 26 December 2022
Chen Y, Fan L, Cai J, Xiang B, Liao M, Xu C, Ren T (2020) Research progress on the difference of pathogenicity of Newcastle disease virus infecting different poultry. Poult Husb Dis Control 9:33–36
Meng L, Pang H, Zhao Z, Zhao K, Yuan W (2021) Status on Newcastle disease vaccines. Prog Vet Med 42:107–111
Li M, Li Y, Yang B, Xu Z, Wang H, Qiu X, Sun Y, Tan L, Liao Y, Song C, Liu W (2020) Identification and whole genome sequencing of a genotype VII Newcastle Disease Virus isolated from pigeon. Chin J Vet Med 56:5–9+14
Selim KM, Selim A, Arafa A, Hussein HA, Elsanousi AA (2018) Molecular characterization of full fusion protein (F) of Newcastle disease virus genotype VIId isolated from Egypt during 2012–2016. Vet World 11:930–938
Dortmans J, Koch G, Rottier PJM, Peeters BPH (2009) Virulence of pigeon paramyxovirus type 1 does not always correlate with the cleavability of its fusion protein. J Gen Virol 90:2746–2750
Dimitrov KM, Ramey AM, Qiu X, Bahl J, Afonso CL (2016) Temporal, geographic, and host distribution of avian paramyxovirus 1(Newcastle disease virus). Infect Genet Evol 39:322–334
Tong Z, Cao L, Cao Y, Wang X, Cheng W, Ye L, Sun H, Sun Y, Guo Y (1998) Report on the diagnosis of a Newcastle disease transmitted from a magpie to chickens. Heilongjiang Anim Sci Vet Med 4:38
Yang J (2002) Magpie Newcastle disease. Chin J Vet Med 3:52
Wu G, Wang Z, Zhang G, Zhou G (2007) Magpie Newcastle disease cases. Chin J Vet Med 5:56
Ministry of Agiculture and Rural Affairs of the People’s Republic of China (2020) GB/T 16550–2020, Diagnostic techniques for Newcastle disease, pp 2–3
National Standards of the People’s Republic of China (2009) Diagnostic techniques for Newcastle disease. Issued by Standardization Administration of the People’s Republic of China
Kaleta EF, Alexander DJ, Russell PH (1985) The first isolation of the avian PMV-1 virus responsible for the current panzootic in pigeons? Avian Pathol 14:553–557
Alexander DJ (2000) Newcastle disease and other avian paramyxovirus. Rev Sci Tech 19:443–462
Saif YM (2012) Diseases of poultry. China Agric Press 12:65–92
Zhao Y, Zhao Y, Wu H, Yang Z, Liu H (2022) Analysis of epidemic genotypes of Newcastle Disease Virus isolated from poultry in China. Prog Vet Med 43:107–111
Luo Y, Wang J, Wang Y, Wei R, Lv Y, Zhao Y, Zheng D, Yu S, Zhang L, Liu H (2018) Distribution and molecular characteristics of 10 strains of pigeon paramyxovirus in some regions of China from 2014 to 2017. Chin J Vet Sci 38:1883–1886+1931
Qiu X, Meng C, Zhan Y, Yu S, Li S, Ren T, Yuan W, Xu S, Sun Y, Tan L, Song C, Liao Y, Ding Z, Liu X, Ding C (2017) Phylogenetic, antigenic and biological characterrizeation of pigeon paramyxovirus type 1 circulating in China. Virol J 14:186
Tian Y, Xue R, Yang W, Li Y, Xue J, Zhang G (2020) Characterization of ten paramyxovirus type 1 viruses isolated from pigeons in China during 1996–2019. Vet Microbiol 244:108661
Li T, Lu B, Ding S, Tang J, Wang C, Liu H (2021) Isolation and identification of an isolate of pigeon Paramyxovirus type 1 and sequence analysis of F gene. Heilongjiang Anim Sci Vet Med 24:51–56
Liu W, Wang X, Liu C, Li C, Tong Z (2003) Diagnostic report of Newcastle disease in chickens caused by magpie Newcastle disease. Heilongjiang Anim Sci Vet Med 8:80
Wang Z, Wu G, Zhang G, Liu J, Zheng C, Lv L (2009) Isolation, identification and biological characteristics of Newcastle disease virus in magpies. Heilongjiang Anim Sci Vet Med 5:92–93
Liu H, Li T, Tang J, Ding S, Lu B, Wang C (2021) Isolation, identification and virulence analysis of NDV isolated from Shanxi Province. Mod J Anim Husb Vet Med 11:43–47
Liu Y, Xue J, Ma X, Wang G, Wang Z, Zhang W (2016) 1 Isolation and identification of a virulent strain of Wu ji Newcastle Disease virus and genetic evolution analysis of F gene. J Econ Anim 20:13–17
Yang S, Huang Q, Cui N, Sun S, Xu C (2022) Molecular characterization and phylogenetic analysis of Newcastle disease virus isolates from healthy wild birds. Chin J Vet Sci 42:452–456
Ren S, Xie X, Wang Y, Tong L, Gao X, Jia Y, Wang H, Fan M, Zhang S, Xiao S, Wang X, Yang Z (2016) Molecular characterization of a Class I Newcastle disease virus strain isolated from a pigeon in China. Avian Pathol 45:408–417
Pei Y, Sun Y, Zhao Y, Zhang G, Xue J (2022) Genome sequencing and pathogenicity analyses of four isolates of the Newcastle Disease Virus in pigeons. Chin J Virol 38:402–414
Olszewska-Tomczyk M, Dolka L, Switon E, Smietanka K (2018) Genetic changes in pigeon paramyxovirus type-1 induced by serial passages in chickens and microscopic lesions caused by the virus in various avian hosts. J Vet Res 62:447–455
Heiden S, Grund C, HÖper D, Mettenleiter TC, RÖmer-OberdÖrfer A, (2014) Pigeon paramyxovirus type l variants with polybasic F protein cleavage site but strikingly different pathogenicity. Virus Genes 49:502–506
Funding
This study was funded by Innovation Team Programes of College of Veterinary Medicine, Shanxi Agricultural University (DY-CX005) and General Program of Shanxi Youth Science and Technology Research Fund (201701D221198).
Author information
Authors and Affiliations
Contributions
Huadong Liu wrote the main manuscript text,All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors declare that they have no conflict of interest.
Research involving human participants and/or animals
All procedures involving animals were approved by the Institutional Animal Care and Use Committee of Shanxi Agricultural University (Permit Number: SXAU-EAW-2022C.AL.00603001) and conducted according to the guidelines for animal welfare by the World Organization for Animal Health.
Additional information
Edited by Nicola Decaro.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, H., Li, T., Ding, S. et al. Complete genome sequence analysis and biological characteristics of Newcastle disease viruses from different hosts in China. Virus Genes 59, 449–456 (2023). https://doi.org/10.1007/s11262-023-01988-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-023-01988-y