Abstract
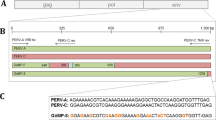
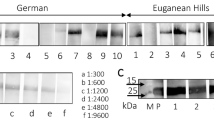
Porcine endogenous retrovirus (PERV), which integrates as a provirus into the genome of pig cells, is an important biosafety issue in xenotransplantation. Screening and analyzing the presence and expression of PERV will provide essential parameters for assessing the biosafety of donor sources. In the present study, we investigated the prevalence of PERV in Diannan small-eared pigs, a unique closed colony that is distributed in southern Yunnan Province in southwestern China. PCR was performed to amplify env-A, env-B, env-C, pol, gag, and mtDNA in peripheral blood samples. The results revealed that PERV env-A, env-B, pol, and gag were detected in all individuals, but env-C was deficient in most pigs, suggesting that the main subtypes of PERVs in Diannan small-eared pigs are PERV-A and PERV-B. Furthermore, PERV pol and the porcine housekeeping gene GAPDH were detected by RT-PCR in all peripheral blood samples, indicating that PERV had transcriptional activity. Finally, the consensus sequences of PERV-A and PERV-B were amplified and digested with KpnI and MboI. Interestingly, a total of seven digestion patterns were obtained, which is less than that observed in other pig breeds. The PCR products were cloned into the pUCm-T vector and sequenced. The results showed that all of the inserts were highly homologous to either PERV-A or PERV-B, and the ratios of PERV-A and PERV-B were 21.1% and 78.9%, respectively. These data suggest that Diannan small-eared pigs may be a candidate donor source for xenotransplantation.




Similar content being viewed by others
References
Cooper DKC, Hara H, Iwase H, Yamamoto T, Li Q, Ezzelarab M, Federzoni E, Dandro A, Ayares D (2019) Justification of specific genetic modifications in pigs for clinical organ xenotransplantation. Xenotransplantation 26(4):e12516
McGregor CGA, Byrne GW (2017) Porcine to human heart transplantation: is clinical application now appropriate? J Immunol Res 2017:1–11
Yamamoto T, Iwase H, King TW, Hara H, Cooper DKC (2018) Skin xenotransplantation: historical review and clinical potential. Burns 44:1738–1749
Li L, Meng H, Zou Q, Zhang J, Cai L, Yang B, Weng J, Lai L, Yang H, Gao Y (2019) Establishment of gene-edited pigs expressing human blood-coagulation factor VII and albumin for bioartificial liver use. J Gastroenterol Hepatol 34(10):1851–1859
Denner J, Scobie L, Schuurman H (2018) Is it currently possible to evaluate the risk posed by PERVs for clinical xenotransplantation? Xenotransplantation 25:e12403
Fishman JA (2018) Infectious disease risks in xenotransplantation. Am J Transpl 18:1857–1864
Łopata K, Wojdas E, Nowak R, Łopata P, Mazurek U (2018) Porcine endogenous retrovirus (PERV): molecular structure and replication strategy in the context of retroviral infection risk of human cells. Front Microbiol 9:730–741
Denner J (2008) Recombinant porcine endogenous retroviruses (PERV-A/C): a new risk for xenotransplantation? Arch Virol 153:1421–1426
Lee D, Lee J, Park N, Oh YK, Kwon M, Kim YB (2008) Analysis of natural recombination in porcine endogenous retrovirus envelope genes. J Microbiol Biotechnol 18:585–590
Denner J (2016) How active are porcine endogenous retroviruses (PERVs)? Viruses 8:215–226
Semaan M, Rotem A, Barkai U, Bornstein S, Denner J (2013) Screening pigs for xenotransplantation: prevalence and expression of porcine endogenous retroviruses in Göttingen minipigs. Xenotransplantation 20:148–156
Xing XW, Hawthorne WJ, Yi S, Simond DM, Dong Q, Ye B, Tong QJ, Ye Z, Wang W (2009) Investigation of porcine endogenous retrovirus in the conservation population of Ningxiang Pig. Transpl Proc 41:4389–4393
Xing XW, Xue LQ, Mo ZH, Huang SQ, Wang W (2006) Porcine endogenous retrovirus in Daweizi pigs in Hunan. J Cent South Univ (Med Sci) 31:838–842
Xing XW, Xu SC, Mo ZH, Ning P, Wang Y, Wang W (2007) Analysis of subtypes of porcine endogenous retrovirus in hybrids of Landrace with Qinghai Bamei swine. Chin J Zoonoses 23:878–882
Le Tissier P (1997) Two sets of human-tropic pig retrovirus. Nature 389(6652):681–682
Takeuchi Y, Patience C, Magre S, Weiss RA, Stoye JP (1999) Host range and interference studies of three classes of pig endogenous retrovirus. J Virol 72(12):9986–9991
Switzer WM, Shanmugam V, Chapman L, Heneine W (1999) Polymerase chain reaction assays for the diagnosis of infection with the porcine endogenous retrovirus and the detection of pig cells in human and nonhuman recipients of pig xenografts. Transplantation 68(2):183–188
Wang S, Wang B, He H, Sun A, Guo C (2018) A new set of reference housekeeping genes for the normalization RT-qPCR data from the intestine of piglets during weaning. PLoS ONE 13:e204583
Lee JH, Webb GC, Allen RD, Moran C (2002) Characterizing and mapping porcine endogenous retroviruses in Westran pigs. J Virol 76:5548–5556
Lu SX, Lian LS (2013) Research progress and development of germplasm resources of diannan small-eared pigs. China Swine Ind 8:165–167
Li B, Zheng H, He BL, Chen LL, Li JT, Jiao JL (2011) Detection of some blood physiological and biochemical parameters in diannan miniature pigs. J Kunming Med Univ 32:13–18
Bosch S, Arnauld C, Jestin A (2000) Study of full-length porcine endogenous retrovirus genomes with envelope gene polymorphism in a specific-pathogen-free Large White swine herd. J Virol 74:8575–8581
Patience C, Takeuchi Y, Weiss RA (1997) Infection of human cells by an endogenous retrovirus of pigs. Nat Med 3:282–286
Martin U, Winkler ME, Id M, Radeke H, Arseniev L, Takeuchi Y, Simon AR, Patience C, Haverich A, Steinhoff G (2000) Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV). Xenotransplantation 7:138–142
van der Laan LJ, Lockey C, Griffeth BC, Frasier FS, Wilson CA, Onions DE, Hering BJ, Long Z, Otto E, Torbett BE, Salomon DR (2000) Infection by porcine endogenous retrovirus after islet xenotransplantation in SCID mice. Nature 407:90–94
Irgang M, Karlas A, Laue C, Specke V, Tacke SJ, Kurth R, Schrezenmeir J, Denner J (2005) Porcine endogenous retroviruses PERV-A and PERV-B infect neither mouse cells in vitro nor SCID mice in vivo. Intervirology 48:167–173
Denner J (2018) Why was PERV not transmitted during preclinical and clinical xenotransplantation trials and after inoculation of animals? Retrovirology 15:28–37
Overbaugh J, Miller AD, Eiden MV (2001) Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol Mol Biol Rev 65:371–389
Dieckhoff B, Petersen B, Kues WA, Kurth R, Niemann H, Denner J (2008) Knockdown of porcine endogenous retrovirus (PERV) expression by PERV-specific shRNA in transgenic pigs. Xenotransplantation 15:36–45
Li ZG, Liu GB, Pan MX, Wu QS, Ge M, Du J, Wang Y, Gao Y (2013) Knockdown of porcine endogenous retroviruses by RNA interference in Chinese experimental miniature pig fibroblasts. Transpl Proc 45:748–755
Demange A, Yajjou-Hamalian H, Gallay K, Luengo C, Beven V, Leroux A, Confort MP, Al AE, Gouet P, Moreau K, Ronfort C, Blanchard Y (2015) Porcine endogenous retrovirus-A/C: biochemical properties of its integrase and susceptibility to raltegravir. J Gen Virol 96:3124–3130
Argaw T, Colon-Moran W, Wilson C (2016) Susceptibility of porcine endogenous retrovirus to anti-retroviral inhibitors. Xenotransplantation 23:151–158
Yang L, Guell M, Niu D, George H, Lesha E, Grishin D, Aach J, Shrock E, Xu W, Poci J, Cortazio R, Wilkinson RA, Fishman JA, Church G (2015) Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 350:1101–1104
Niu D, Wei HJ, Lin L, George H, Wang T, Lee IH, Zhao HY, Wang Y, Kan Y, Shrock E, Lesha E, Wang G, Luo Y, Qing Y, Jiao D, Zhao H, Zhou X, Wang S, Wei H, Guell M, Church GM, Yang L (2017) Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 357:1303–1307
Godehardt AW, Fischer N, Rauch P, Gulich B, Boller K, Church GM, Tönjes RR (2019) Characterization of porcine endogenous retrovirus particles released by the CRISPR/Cas9 inactivated cell line PK15 clone 15. Xenotransplantation 31:e12563
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81560140). We are grateful for the samples provided by the Animal Experimental Center of Kunming Medical University.
Author information
Authors and Affiliations
Contributions
Ruhong Li and Gang Liu designed the study. Yunfei Zhang, Xiaowei Xing, and Peng Li performed the study. Yunfei Zhang, Xiaowei Xing, Lihua Huang, and Yong Wu analyzed the data and wrote the manuscript. All the authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This study was approved by the Experimental Animals Ethics Committee of the Third Xiangya Hospital, Central South University.
Additional information
Edited by Juergen A Richt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Y., Xing, X., Huang, L. et al. Screening pigs for xenotransplantation in China: investigation of porcine endogenous retrovirus in Diannan small-eared pigs. Virus Genes 56, 202–208 (2020). https://doi.org/10.1007/s11262-019-01722-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-019-01722-7