Abstract
Reports of rotavirus excretion in calves usually result from cross-sectional studies, and in face of the conflicting results regarding protection of calves born to vaccinated dams against diarrhea, the aim of the present study was to evaluate rotavirus excretion in dairy calves born to vaccinated or unvaccinated dams, to identify the genotypes of bovine rotavirus group A (RVA) strains isolated from these animals as well as to investigate characteristics of the disease in naturally occurring circumstances throughout the first month of life. Five hundred fifty-two fecal samples were taken from 56 calves, 28 from each farm and, in the vaccinated herd, 11/281 samples (3.91%) taken from six different calves tested positive for RVA while in the unvaccinated herd, 3/271 samples (1.11%) taken from 3 different calves tested positive. The genotyping of the VP7 genes showed 91.2% nucleotide sequence identity to G6 genotype (NCDV strain), and for the VP4 gene, strains from the vaccinated herd were 96.6% related to B223 strain, while strains from the unvaccinated herd were 88% related to P[5] genotype (UK strain). Genotypes found in this study were G6P[11] in the vaccinated herd and G6P[5] in the unvaccinated herd. All calves infected with rotavirus presented an episode of diarrhea in the first month of life, and the discrepancy between the genotypes found in the commercial vaccine (G6P[1] and G10P[11]) and the rotavirus strains circulating in both vaccinated and unvaccinated herds show the importance of keeping constant surveillance in order to avoid potential causes of vaccination failure.
Similar content being viewed by others
Introduction
Neonatal calf diarrhea (NCD) is one of the most important causes of morbidity and mortality in animals aged less than 6 weeks of life, leading to economic losses due to the cost of treatment, prophylaxis, increased susceptibility to other infections, reduced growth rates, and death of calves. Colostrum management, size of the herd, type of housing, nutritional status, and the presence of other pathogens are considered risk factors for the development of the infection (Lorenz et al. 2011; Cho and Yoon 2014).
NCD is a multifactorial disease, associated with infectious and non-infectious causes. Of the bacteria, viruses, and protozoa commonly involved in the etiology of NCD, the most prevalent viral agent is bovine rotavirus group A (RVA), as reported in studies conducted in Brazil, Netherlands, Australia, USA, and New Zealand (Langoni et al. 2004; Bartels et al. 2010; Izzo et al. 2011; Cho et al. 2013; Cho and Yoon 2014; Al Mawly et al., 2015).
Rotavirus is a genus from the Reoviridae family and is divided into 8 groups or species (A–H) based on antigenic properties and nucleotide sequence diversity of the inner viral capsid protein VP6 (Matthijnssens et al. 2012). From these, RVA is the most prevalent (Papp et al. 2013). The non-enveloped virion has a triple-layered capsid surrounding 11 dsRNA segments that encode 6 structural (VP1–4, VP6, and VP7) and 5 non-structural (NSP1-NSP5/6) proteins (Attoui et al. 2011). Since VP4 and VP7 induce the production of neutralizing antibodies and protective immunity, these proteins play a central role in strain selection for vaccine production and form the basis of a binary classification system, namely, the G (glycosylated) and P (protease-sensitive) genotypes (Estes and Greenberg 2013). To date, 27 G and 38 P genotypes with different combinations have been described (Matthijnssens et al. 2011; Trojnar et al. 2013). Regarding bovine rotavirus infections, G6, G8, and G10 together with P[1], P[5], and P[11], respectively, are the G and P genotypes most commonly found (Alkan et al. 2010).
General recommendations regarding decrease of RVA diarrhea include management practices, especially good hygiene and sanitation procedures, as well as pathogen-specific interventions, such as the use of vaccine prophylaxis (Alkan et al. 2010; Izzo et al. 2011). The vaccination of cows aims an increase of the concentration of specific antibodies in colostrum against targeted pathogens (Kaplon et al. 2013). Although many reports state that vaccination programs of pregnant cows are one of the main strategies for prevention of RVA diarrhea in calves, it must be kept in mind that calves that receive a sufficient quantity of specific antibodies through colostrum intake are not completely protected against rotavirus infection, instead, the severity of clinical signs is decreased, reducing deaths from RVA infection (Parreño et al. 2004; Kaplon et al. 2013).
The aim of this study was to evaluate rotavirus excretion and to identify the genotypes of RVA strains isolated from a vaccinated and an unvaccinated dairy herd from São Paulo state, Brazil.
Materials and methods
Fifty-six calves were evaluated in a longitudinal design study. These animals were allotted in two experimental groups with 28 animals: group V comprised of animals that belonged to a herd that regularly vaccinated the dams with a commercial vaccine against NCD, and group NV comprised of animals from a herd that did not vaccinate the dams against NCD. The vaccination scheme included administration of a commercial adjuvant vaccine containing inactivated bovine RVA strains G6P[1] and G10P[11], inactivated bovine coronavirus, Escherichia coli K99 colibacterin and type C Clostridium perfringens toxoid according to manufacturer’s instructions—pregnant cows received two intramuscular doses of vaccine, with an interval of approximately 3 weeks, the second dose being administered 3 to 6 weeks pre-partum. Cows from group NV did not receive the vaccine in the prepartum period.
Besides vaccination management, herd V comprised 1400 Holstein cows in lactation, with a mean production of 32 L of milk/cow/day, and group NV comprised 100 Holstein cows in lactation, with a mean production of 20 L of milk/cow/day. Colostrum management was similar in both farms, and only calves born to cows that produced a sufficient volume of colostrum to ensure the intake of 4 L in the first meal were included in this study. Colostrum management consisted of bottle feeding calves ad libitum with their own mother’s colostrum up to 2 h after birth, with a minimum volume of colostrum intake of 4 L per calf.
The fecal samples were collected in two municipalities located in the state of São Paulo in 2011 and 2012. Holstein calves were randomly selected and fecal samples were taken directly from the rectum using individual plastic bags before colostrum intake, and at 1, 2, 7, 15, 21, and 30 days of age and frozen at −20 °C until analysis. Whenever calves presented diarrhea, samples were taken at 24 h intervals until animals presented two consecutive days of normal feces.
Fecal samples were classified in scores according to consistency: score 0 (firm feces), score 1 (slightly loose feces), score 2 (loose feces), and score 3 (watery feces) (Fernandez et al., 1998). Scores 0 and 1 were considered normal feces and scores 2 and 3 were considered diarrhea.
Samples were screened for RVA by silver stained-polyacrylamide gel electrophoresis (PAGE) technique (Herring et al. 1982) modified by Pereira et al. (1983), which is a technique with sensitivity approximately equal to electron microscopy (Herring et al. 1982).
The RT-PCR was employed to characterize the bovine RVA genotypes G and P as described previously (Silva et al. 2015). Fecal samples were prepared as 50% (v/v) suspensions in DEPC-treated ultrapure water and centrifuged at 5000×g for 10 min at 4 °C. Total RNA was extracted from 250 μL of the supernatant using TRIzol Reagent™ (Invitrogen, Carlsbad, CA, USA) and cDNA was synthesized using random primers (Invitrogen, Carlsbad, CA, USA) and SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) as per manufacturer’s protocol. Different PCR runs were performed using primers Beg9, End9, End9CRW8, End9UK (for G genotypes) or con3 and con2 (for P genotypes) (Gouvea et al. 1994a; Gouvea et al. 1994b; Gentsch et al. 1992; Silva et al. 2015). Therefore, 2.5 μL cDNA was added into the reaction mix comprising 1× PCR Buffer™ (Invitrogen, Carlsbad, CA, USA), 0.2 mM of each dNTP, 0.5 μM of each forward and reverse primers, 1.5 mM MgCl2, and 3 U Platinum Taq Polymerase™ (Invitrogen, Carlsbad, CA, USA) and nuclease-free water to obtain a final volume of 25 μL. This solution was then heated to 94 °C for 3 min, followed by 40 amplification cycles, with 1 cycle consisting of 1 min at 94 °C, 1.5 min at 50 °C, and 1 min at 72 °C; and a final extension step of 10 min at 72 °C. After completion of thermocycles, PCR products were separated by electrophoresis on a 1.5% agarose gel, stained with ethidium bromide, and visualized by UV transillumination.
Ten samples in which RT-PCR results were satisfactory were used for genotyping. The amplicons were purified using ExoSAP-IT® PCR product cleanup (USB Products Affymetrix, Cleveland, USA) and nucleotide sequences of the VP4 and VP7 gene segments were determined in both directions using BigDye Terminator 3.1™ cycle sequencing kit (Applied Biosystems, Foster City, USA), according to the manufacturer’s protocol and a 3500™ Genetic Analyzer system (Applied Biosystems, Foster City, USA). The nucleotide sequences were assembled using Bioedit 7.0.5 software (Hall 1999) and analyzed using the RotaC 2.0 automated genotyping tool for Group A rotavirus to confirm the RVA identity of all sequences (Maes et al. 2009). Phylogenetic trees were constructed and molecular analyses were performed using the MEGA® 6.06 software by using the neighbor-joining algorithm and the maximum composite likelihood DNA substitution model for each viral gene or nucleotide fragment (Tamura et al. 2013). Statistical support was obtained by 1000 bootstrap replicates. The identity matrix was constructed using Bioedit software version 7.0.5. Sequences from this study were deposited in the GenBank database under the following accession numbers: VP7 (KX121165, KX121166, KX121167, KX121168, KX121169, KX121170, KX121171, KX121172, KX121173, KX121174) and VP4 (KX121175, KX121176, KX121177, KX121178, KX121179, KX121180, KX121181, KX121182, KX121183, KX121184).
Results
The number of fecal samples taken from calves from the vaccinated and unvaccinated herds; its distribution by scores as well as the number of bovine RVA positive samples according to fecal score are shown in Table 1.
Fecal samples positive for rotavirus detection through PAGE, number and age of calves that tested positive for RVA in feces by this technique, number of days of elimination of diarrheic feces throughout the first month of life, as well as results of genotyping are shown in Table 2. The distribution pattern of the 11 dsRNA segments was 4:2:3:2 in all the positive samples by PAGE technique.
All 14 samples analyzed by RT-PCR for the detection of VP4 were positive. Regarding VP7, in four samples from three different animals, it was not possible to obtain amplicons using the primers described, even though samples were tested twice, in different moments, using new RNA extractions, and positive and negative controls worked as expected. As sequencing was performed in only ten samples in which RT-PCR results were adequate, including at least one sample from each positive calf, the genotype of the four samples aforementioned could not be determined.
Genotyping revealed that all RVA positive calves from group V were infected with G6P[11] RVA, while in group NV, the genotype found was G6P[5]. Also, G6P[11] strain was detected in both normal and diarrheic feces, while G6P[5] strain was found only in diarrheic feces. Positive calves excreted the virus for 1 to 3 days in the vaccinated herd, and for only 1 day in the unvaccinated herd (Table 2).
None of the calves that tested positive for RVA in feces were aged between 1 and 7 days of life. Calves that tested positive for rotavirus in feces were in their second (n = 4), third (n = 4), or fourth (n = 1) weeks of age.
Evaluation of fecal scores of calves in the first 30 days of life revealed that in group V, 72.6% of fecal samples taken were normal, while 27.4% of feces were diarrheic. In group NV, 72.7% of fecal samples were normal while 27.3% were diarrheic.
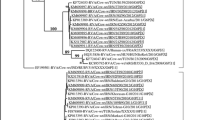
The nucleotide sequence analysis of the VP7 genes of 10 Brazilian bovine RVA strains belonging to both the G6P[11] and G6P[5] genotypes showed the highest nucleotide sequence identity (91.2%) to G6 genotype (NCDV strain) (Fig. 1). For the VP4 gene, two Brazilian bovine RVA strains (THAF16 and THAF22) were most closely related to P[5] genotype (UK strain) (88%). Furthermore, the VP4 gene of the remaining Brazilian bovine strains belonging to genotype G6P[11] from this study was closely related to B223 strain (96.6%) (Fig. 2).
Maximum likelihood phylogenetic tree based on the partial ORF nucleotide sequences of bovine RVA VP4 gene. The bovine strains analyzed in this study are marked with a lozange. The scale bar indicates nucleotide substitutions per site. Bootstrap values (1000 replicates) above 70 are shown in the corresponding nodes
Maximum likelihood phylogenetic tree based on the partial ORF nucleotide sequences of bovine RVA VP7 gene. The bovine strains analyzed in this study are marked with a lozange. The scale bar indicates nucleotide substitutions per site. Bootstrap values (1000 replicates) above 70 are shown in the corresponding nodes
Discussion
Although positive samples were found in both normal (n = 5) and diarrheic feces (n = 9), which agrees with the report by Bartels et al. (2010) and Freitas et al. (2011), all calves that tested positive for rotavirus detection in feces presented an episode of diarrhea in the first 30 days of life. This finding is relevant since most of the cross-sectional field research performed investigating RVA diarrhea, consider calves in which RVA is found in normal feces to be asymptomatic. In the report by Oliveira-Filho et al. (2007) and Cho et al. (2013), bovine RVA was only found in diarrheic feces, however, this sort of information is rare in the literature consulted, which can lead to misinformation. It is quite likely that newborn calves infected with RVA will always manifest an episode of diarrhea, and that the virus can be eliminated in all stages of clinical course of the disease, including in moments in which feces are normal. Thus, it would be relevant to perform more longitudinal studies in which fecal samples are taken in predetermined moments, as well as whenever the calves manifest diarrhea, in order to evaluate a larger number of animals to confirm these findings and if they are related to the virulence of genotypes of RVA.
Nourmohammadzadeh et al. (2012) reported that the higher prevalence of rotavirus infection occurs in calves aged between 2 and 4 weeks, and lower in animals in the first week of life, which agrees with the results of the present report, in which none of the positive calves were in their first week of life, four calves were in the second week of life, four calves were in the third week of life and one calf was in the fourth week of life.
Bartels et al. (2010), evaluating the prevalence and risk factors for the presence of enteropathogens in normal and diarrheic feces of 424 calves aged between 1 and 3 weeks of life reported that the prevalence of normal consistency feces was 57.1%, semiliquid feces was 23.8%, and liquid feces was 19.1%. In the current study, considering the same age groups of calves, percentages found were 72.3 and 71% of normal consistency feces, 19.4 and 19% of semiliquid feces, and 8.30 and 10% of liquid feces, in groups V and NV, respectively. These authors also reported that the percentage of calves with diarrheic feces in the second week of life was higher than in calves with 1 or 3 weeks of age, which was similar to the findings of the present study, in which the percentage of diarrheic calves in their second week of life was 60.7%. This finding is attributable to the immune status of calves, which is higher in the first week of life due to passive immunity provided by colostrum, and gradually decreases through a natural process of catabolism of immunoglobulins, making calves more susceptible to infection in the second and third weeks of life. Since most cases of RVA diarrhea occur within these age groups, at 4 weeks of age, calves are normally immune to infection through natural resistance (Chase et al. 2008; Coura et al. 2015).
The duration of rotavirus excretion is variable among studies found in literature. A recent report of longitudinal evaluation of diarrhea occurrence in a dairy herd in Brazil stated that the excretion of rotavirus in feces lasted for 1–3 days, which agrees with the results of the present study (Coura et al. 2015). In experimental studies, rotavirus excretion has been reported to last up to 10 days (Fernandez et al. 1998). This difference might be attributable to the method of detection of RVA in feces, since both field studies employed PAGE as a screening method for rotavirus in feces, and the limitation of this method lies in the need for a certain concentration of virus particles for the test to be positive, while in the experimental study cited, the method used (ELISA) is considered a test able to detect smaller amounts of virus present in feces than PAGE.
It is important to point out that there was a difference between the RVA strain found between the vaccinated calves (G6P[11]) and the strains present in the commercial vaccine (G6P[1], and G10P[11]), which might be one of the contributing factors to the lack of efficiency in protecting calves of group V from rotavirus diarrhea. According to Alkan et al. (2010), several factors can contribute to vaccine failures, especially vaccination management. In the present study, there was a special attention with the conservation and protocol of vaccination employed, in order to avoid the risk of vaccine failure due to inappropriate conservation of the vaccine or a longer period of time between the second dose of vaccine and parturition. However, contrary to expectations, incidence of rotavirus diarrhea was higher in group V than in group NV, which might be due to differences in herd size, exposure to virus, and management procedures between herds (Meganck et al. 2015).
The strain found in the vaccinated herd (G6P[11]) was different from the strain of the non-vaccinated herd (G6P[5]), and none of these strains were present in the commercial vaccines available by the time of the experiment. In Brazil, G6P[11] has been incriminated as a cause of a diarrhea outbreak in a beef herd (Medeiros et al. 2014a), although there are also reports of its occurrence as the most common genotypic combination in some states of country (Silva et al. 2012). In Ireland, G6P[11] is reported as a low prevalence genotypic combination found especially in dairy herds (Collins et al. 2014), while in Turkey, Alkan et al. (2010) reported a high prevalence of this genotype in either unvaccinated herds and in herds immunized against G6P[1].
Vaccines containing G6P[1] provide poor heterologous protection against G6 bovine RVA that contain different P genotypes from the vaccine strains (Alkan et al. 2010; Medeiros et al. 2014b). This implies that, considering the latest surveillances performed in Brazil and in other regions of the world, strains of RVA present in the commercial vaccines should be replaced by genotypic combinations more prevalent in the region of interest to reduce the occurrence of RVA diarrhea and all the economic losses associated to this infirmity.
In a recent review performed by Papp et al. (2013) on the RVA strains reported in cattle worldwide, the most prevalent genotypic combinations found in diarrheic calves were G6P[5] (UK strain), G6P[11] (KN4 strain), and G10P[11] (B223 strain), comprising 40.4% of all RVA strains. Also, in Argentina, the most prevalent genotypic combinations reported were G6P[11] in dairy herds and G6P[5] in beef herds (Badaracco et al. 2013).
Parreño et al. (2004) reported that the protective effect of high titers of serum antibodies against bovine RVA significantly reduces the occurrence of diarrhea caused by rotavirus, but does not eliminate the virus from the herd, which reinforces the concept that commercial vaccines currently used are not designed to prevent rotavirus infection, but rather to decrease morbidity, severity of diarrhea, and mortality in field conditions. Although this might have impact in some of the economic costs associated with neonatal calf diarrhea, it does not reduce the expenses of treatment aiming to avoid secondary bacterial infections and might still impact on the potential development and performance of calves in their productive life, given that all the animals positive for RVA detection in the present study manifested at least one episode of diarrhea throughout the first month of life.
The nucleotide sequence analysis of the VP7 genes of bovine RVA strains showed 91.2% nucleotide sequence identity to NCDV strain (G6 genotype). In the unvaccinated herd, the VP4 gene was most closely related (88%) to P[5] genotype (UK strain), while in the vaccinated herd, the VP4 gene detected was 96.6% related to the B223 strain. Alkan et al. (2010) reported that the G6P[11] lineage found in Turkey formed a unique G6 lineage with only 90.1% amino acid identity to the G6 strain NCDV found in the vaccines. Medeiros et al. (2014a) also reported an outbreak of diarrhea in beef calves in Brazil caused by a G6P[11] distant from the most common lineages of RVA strains G6P[1] and G6P[5]. As the strain found in the vaccinated herd of the present study showed a nucleotide sequence identity of 91.2% to the vaccinal strain NCDV, and the VP4 antigen associated was different from the vaccinal strain, this could help to explain the lack of protection of the vaccinated herd against RVA diarrhea.
The small number of calves positive to RVA infection found in our study, especially in the unvaccinated herd, as well as the impossibility of comparing herds with similar sizes, production and infection pressure constituted the main limitations of the study, although the results presented here provided important highlights to the characteristics of natural occurring rotavirus diarrhea in calves in the first month of life. More field studies should be performed to evaluate the genotypic profile of RVA present in both vaccinated and unvaccinated herds.
The results of this study show that all calves infected with rotavirus presented an episode of diarrhea in the first month of life, and that the discrepancy between the genotypes found in the commercial vaccine and the rotavirus strains circulating in both vaccinated and unvaccinated herds, once more, reinforces the importance of constant surveillance in order to avoid potential causes of vaccination failure.
References
Al Mawly, J., Grinberg, A., Prattley, D., Moffat, J., and French, N., 2015. Prevalence of endemic enteropathogens of calves in New Zealand dairy farms, New Zealand Veterinary Journal, 63, 147--152
Alkan, F., Ozkul, A., Oguzoglu, T.C., Timurkan, M.O., Caliskan, E., Martella, V., and Burgu, I., 2010. Distribution of G (VP7) and P (VP4) genotypes of group A rotaviruses from Turkish calves with diarrhea, 1997-2008, Veterinary Microbiology, 141, 231--237
Attoui, H., Mertens, P.P.C., Becnel, J., Belaganahalli, S., Bergoin, M., Brussaard, C.P., Chappell, J.D., Ciarlet, M., and del Vas, M., 2011. Family reoviridae–Virus Taxonomy Ninth Report of the International Committee on Taxonomy of Viruses, (Elsevier Academic Press, Amsterdam)
Badaracco, A., Garaicochea, L., Matthijnssens, J., Louge Uriarte, E., Odeón, A., Bilbao, G., Fernandez, F., Parra, G.I., and Parreño, V., 2013. Phylogenetic analyses of typical bovine rotavirus genotypes G6, G10, P[5] and P[11] circulating in Argentinean beef and dairy herds, Infection, Genetics and Evolution, 18, 18--30
Bartels, C.J.M., Holzhauer, M., Jorritsma, R., Swart, W.A.J.M., and Lam, T.J.G.M., 2010. Prevalence, prediction and risk factors of enteropathogens in normal and non-normal faeces of young Dutch dairy calves, Preventive Veterinary Medicine, 93, 162--169
Chase, C.C.L., Hurley, D.J., and Reber, A.J., 2008. Neonatal immune development in the calf and its impact on vaccine response, Veterinary Clinics of North America: Food Animal Practice, 24, 87--104
Cho, Y.I., and Yoon, K.J., 2014. An overview of calf diarrhea—infectious etiology, diagnosis, and intervention, Journal of Veterinary Sciences, 15, 1--17
Cho, Y.I., Han, J.I., Wang, C., Cooper, V., Schwartz, K., Engelken, T., and Yoon, K.Y., 2013. Case-control study of microbiological etiology associated with calf diarrhea, Veterinary Microbiology, 166, 375--385
Collins, P.J., Mulherin, E., Cashman, O., Lennon, G., Gunn, L., O’Shea, H., and Fanning, S., 2014. Detection and characterisation of bovine rotavirus in Ireland from 2006–2008, Irish Veterinary Journal, 67:13
Coura, F.M., Freitas, M.D., Ribeiro, J., Leme, R.A., Souza, C., Alfieri, A.A., Facury Filho, E.J., Carvalho, A.U., Silva, M.X., Lage, A.P., and Heinemann, M.B., 2015. Longitudinal study of Salmonella spp., diarrheagenic Escherichia coli, Rotavirus, and Coronavirus isolated from healthy and diarrheic calves in a Brazilian dairy herd, Tropical Animal Health and Production, 47, 3--1
Estes, M., and Greenberg, H.B., 2013. Rotaviruses. In: Knipe, D.M., Howley, P.M., Cohen, J.I., Griffin, D.E., Lamb, R.A., Martin, M.A., Roizman, B., and Racaniello, V.R. (Eds.), Fields Virology, (Lippincott Williams and Wilkins, Philadelphia)
Fernandez, F.M., Conner, M.E., Hodgins, D.C., Parwani, A.V., Nielsen, P.R., Crawford, S.E., Estes, M.K., and Saif, L.J., 1998. Passive immunity to bovine rotavirus in newborn calves fed colostrum supplements from cows immunized with recombinant SA11 rotavirus core-like particle (CLP) or virus-like particle (VLP) vaccines, Vaccine, 16, 507--516
Freitas, P.P.S., Uyemura, S.A., Silva, D.G., Samara, S.I., and Buzinaro, M.G., 2011. Rotavirus in cattle: risk factors, prevalence and antigenic characterization from dairy calves’ samples in São Paulo State, Brazil, Arquivo Brasileiro de Medicina Veterinária e Zootecnia, 63, 820--827
Gentsch, J.R., Glass, R.I., Woods, P., Gouvea, V., Gorziglia, M., Flores, J., Das, B.K., and Bhan, M.K., 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction, Journal of Clinical Microbiology, 30, 1365--1373
Gouvea, V., Santos, N., and Timenetsky, M.C. 1994a. Identification of bovine and porcine rotavirus G types by PCR, Journal of Clinical Microbiology, 32, 1338--1340
Gouvea, V., Santos, N., and Timenetsky, M.C. 1994b. VP4 typing of bovine and porcine group A rotaviruses by PCR, Journal of Clinical Microbiology, 32, 1333--1337
Hall, T.A., 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95--98
Herring, A.J., Inglis, N.F., Ojeh, C.K., Snodgrass, D.R., and Menzies, J.D., 1982. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels, Journal of Clinical Microbiology, 16, 473--77
Izzo, M.M., Kirkland, P.D., Mohler, V.L., Perkins, N.R., Gunn, A.A., and House, J.K., 2011. Prevalence of major enteric pathogens in Australian dairy calves with diarrhea, Australian Veterinary Journal, 89, 167--173
Kaplon, J., Fremy, C., Bernard, S., Rehby, L., Aho, S., Pothier, P., Ambert-Balay, K., 2013. Impact of rotavirus vaccine on rotavirus genotypes and caliciviruses circulatingin French cattle, Vaccine, 31, 2433--2440
Langoni, H., Linhares, A.C., Avila, F.A., Da Silva, A.V., and Elias, A.O., 2004. Contribution to the study of diarrhea etiology in neonate dairy calves in São Paulo state, Brazil, Brazilian Journal of Veterinary Research and Animal Science, 41, 313--319
Lorenz, I., Fagan, J., and More, S.J., 2011. Calf health from birth to weaning: II. Management of diarrhea in pre-weaned calves, Irish Veterinary Journal, 64, 9
Maes, P., Matthijnssens, J., Rahman, M., and Van R.M., 2009. RotaC: a web-based tool for the complete genome classification of group A rotaviruses, BMC Microbiology, 9, 238
Matthijnssens, J., Ciarlet, M., McDonald, S.M., Attoui, H., Banyai, K., Brister, J.R., Buesa, J., Esona, M.D., Estes, M.K., Gentsch, J.R., Iturriza-Gómara, M., Johne, R., Kirkwood, C.D., Martella, V., Mertens, P.P.C., Nakagomi, O., Parreño, V., Rahman, M., Ruggeri, F.M., Saif, L.J., Santos, N., Steyer, A., Taniguchi, K., Patton, J.T., Desselberger, U., and Van Ranst, M., 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG), Archives of Virology, 156, 1397--1413
Matthijnssens, J., Otto, P.H., Ciarlet, M., Desselberger, U., Van Ranst, M., and Johne, R., 2012. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation, Archives of Virology, 157, 1177--1182
Medeiros, T.N.S., Lorenzetti, E., Alfieri, A.F., and Alfieri, A.A., 2014a. Severe diarrhea outbreak in beef calves (Bos indicus) caused by G6P[11], an emergent genotype of bovine rotavirus group A, Pesquisa Veterinária Brasileira, 34, 717--722
Medeiros, T.N.S., Lorenzetti, E., Alfieri, A.F., and Alfieri, A.A., 2014b. Phylogenetic analysis of a G6P[5] bovine rotavirus strain isolated in a neonatal diarrhea outbreak in a beef cattle herd vaccinates with G6P[1] and G10P[11] genotypes, Archives of Virology, 160, 447--451
Meganck, V., Hoflack, G., Piepers, S., and Opsomer, G., 2015. Evaluation of a protocol to reduce the incidence of neonatal calf diarrhoea on dairy herds, Preventive Veterinary Medicine, 118, 64--70
Nourmohammadzadeh, F., Davoudi, Y., Abdollahpour, G., and Nouri, A., 2012. The prevalence of rotavirus in neonatal calf diarrhea, using electron microscopic examination, Comparative Clinical Pathology, 21, 1231--1234
Oliveira-Filho, J.P., Silva, D.P.G., Pacheco, M.D., Mascarini, L.M., Ribeiro, M.G., Alfieri, A.A., Alfieri, A.F., Stipp, D.T., Barros, B.J., and Borges, A.S., 2007. Diarrhea in Nellore calves: clinical and etiological study, Pesquisa Veterinária Brasileira, 27, 419--424
Papp, H., Lászlá, B., Jakab, F., Ganesh, B., De Grazia, S., Matthijnssens, J., Ciarlet, M., Martella, V., and Bányai, K., 2013. Review of group A rotavirus strains reported in swine and cattle, Veterinary Microbiology, 165, 190--199
Parreño, V., Béjar, C., Vagnozzi, A., Barrandeguy, M., Costantini, V., Craig, M.I., Yuan, L., Hodgins, D., Saif, L.J., and Fernández, F., 2004. Modulation by colostrum-acquired maternal antibodies of systemic and mucosal antibody responses to rotavirus in calves experimentally challenged with bovine rotavirus, Veterinary Immunology and Immunopathology, 100, 7--24
Pereira, H.G., Azeredo, R.S., Leite, J.P., Candeias, J.A., Rácz, M.L., Linhares, A.C., Gabbay, Y.B., and Trabulsi, J.R., 1983. Electrophoretic study of the genome of human rotaviruses from Rio de Janeiro, São Paulo and Pará, Brazil, Journal of Hygiene, 90, 117–125
Silva, F.D.F., Gregori, F., Gonçalves, A.C.S., Samara, S.I., and Buzinaro, M.G., 2012. Molecular characterization of group A bovine rotavirus in southeastern and central-western Brazil, 2009-2010, Pesquisa Veterinária Brasileira, 32, 237--242
Silva, F.D.F., Espinosa, L.R.L., Tonietti, P.O., Barbosa, B.R.P., and Gregori, F., 2015. Whole-genomic analysis of 12 porcine group A rotaviruses isolated from symptomatic piglets in Brazil during the years of 2012–2013, Infection, Genetics and Evolution, 32, 239--254
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar S., 2013. MEGA6: molecular evolutionary genetics analysis version 6.0, Molecular Biology and Evolution, 30, 2725--2729
Trojnar, E., Sachsenröder, J., Twardziok, S., Reetz, J., Otto, P.H., and Johne, R., 2013. Identification of an avian group A rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses, Journal of General Virology, 94, 136--142
Acknowledgements
This study was supported by São Paulo Research Foundation (FAPESP).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Statement of animal rights
The experimental design was approved by the Animal Research Ethics Committee of the School of Veterinarian Sciences of São Paulo State University (Unesp), under protocol number 015110/10.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Rocha, T.G., Silva, F.D.F., Gregori, F. et al. Longitudinal study of bovine rotavirus group A in newborn calves from vaccinated and unvaccinated dairy herds. Trop Anim Health Prod 49, 783–790 (2017). https://doi.org/10.1007/s11250-017-1263-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-017-1263-2