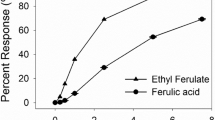
Abstract
Antifungal compounds are of interest to reduce commodity spoilage and exposure to mycotoxins. In this study, a series of quantitative structure-activity relationship (QSAR) equations based on topological properties were developed to gain insight into the antifungal activities of phenolic compounds. The molecules were geometry optimized using B3LYP/6-311+G** density functional theory calculations. Analysis of the frontier orbital properties revealed that conjugated phenolic compounds possessed smaller band gap energies. Genetic function approximation (GFA) on populations of 100 one to two descriptor models over 10,000 generations identified several models for antifungal activity against Fusarium verticillioides, Fusarium oxysporum, Aspergillus flavus, Aspergillus fumigatus, Penicillium expansum, and Penicillium brevicompactum. Phenolic compounds with greater antifungal activity possessed a lower electrophilicity index. The correlation coefficients for the one and two descriptor models ranged from 0.627 to 0.790 and 0.762 to 0.939, respectively. Molecular descriptors associated with electrostatic and topological properties are important to describe the antifungal activities of the phenolic compounds studied.




Similar content being viewed by others
References
Marchese A, Orhan IE, Daglia M, Barbieri R, Di Lorenzo A, Nabavi SF, Gortzi O, Izadi M, Nabavi SM (2016). Food Chem 210:402
Zeidan R, Ul-Hassan Z, Al-Thani R, Balmas V, Jaoua S (2018). Toxins 10:242
Chandra H, Bishnoi P, Yadav A, Patni B, Mishra AP, Nautiyal AR (2017). Plants 6:16
Arnold WA, Oueis Y, O’Connor M, Rinaman JE, Taggart MG, McCarthy RE, Foster KA, Latch DE (2017). Environ Sci: Processes Impacts 19:324
Hartung T (2019) EFSA Journal 17 (S1). DOI. https://doi.org/10.2903/j.efsa.2019.e170710
Aouidate A, Ghaleb A, Ghamali M, Chtita S, Ousaa A, Choukrad M, Sbai A, Bouachrine M, Lakhlifi T (2018). Struct Chem 29:1287
Boberg J, Dybdahl M, Petersen A, Hass U, Svingen T, Vinggaard AM (2019). Curr Opin Toxicol 15:1
Guenard R (2019). Inform 30:6
Tcheremenskaia O, Battistelli CL, Giuliani A, Benigni R, Bossa C (2019). Comput Toxicol 11:91
Shao Y et al (2006). Phys Chem Chem Phys 8:3172
Hypercube, Inc. (2006) Gainesville, FL
Hong H et al (2008). J Chem Inf Model 48:1337
de Oliveira DB, Gaudio AC (2000). Quant Struct-Act Relat 19:599
Zabka M, Pavela R (2013). Chemosphere 93:1051
Dambolena JS, Lopez AG, Meriles JM, Rubinstein HR, Zygadlo JA (2012). Food Control 28:163
Koopmans T (1934). Physica 1:104
Appell M, Bosma WB (2015). J Hazard Mater 288:113
Parr RG, Szentpaly LV, Liu S (1999). J Am Chem Soc 121:1922
Gramatica P, Chirico N, Papa E, Cassani S, Kovarich S (2013). J Comput Chem 34:2121
Gramatica P, Cassani S, Chirico N (2014). J Comput Chem 35:1036
QSAR Research Unit in Environmental Chemistry and Ecotoxicology. http://www.qsar.it Accessed 23 April, 2020
Roy K, Das RN, Ambure P, Aher RB (2016). Chemom Intell Lab Syst 152:18
Gramatica P, Sangion A (2016). J Chem Inf Model 56:1127
Compton DL, Goodell JR, Grall S, Evans KO, Cermak SC (2015). Ind Crop Prod 77:787
Kikuzaki H, Hisamoto M, Hirose K, Akiyama K, Taniguchi H (2002). J Agric Food Chem 50:2161
Hernández-García L, Sandoval-Lira J, Rosete-Luna S, Niño-Medina G, Sanchez M (2018). Struct Chem 29:1265
Ruhland CT, Day TA (2000). Physiol Plant 109:244
Rozema J, Noordijk AJ, Broekman RA, van Beem A, Meijkamp BM, de Bakker NVJ, van de Staaij JWM, Stroetenga M, Bohncke SJP, Konert M, Kars S, Peat H, Smith RIL, Convey P (2001). Plant Ecol 154:9
Kim KH, Chang SW, Lee KR (2010). Can J Chem 88:519
Zhou S, Zhang YK, Kremling KA, Ding Y, Bennett JS, Bae JS, Kim DK, Ackerman HH, Kolomiets MV, Schmelz EA, Schroeder FC, Buckler ES, Jande G (2019). New Phytol 221:2096
Dowd PF, Berhow MA, Johnson ET (2018). Plant Gene 13:50
Kuo Y-H, Hsu Y-W, Liaw C-C, Lee JK, Huang H-C, Kuo L-MY (2005). J Nat Prod 68:1475
Compton DL, Laszlo JA (2002) Novel sunscreens from vegetable oil and plant phenols. US Patent 6:346,236
DeFilippi LJ, Grall SG, Kinney JF, Laszlo JA, Compton DL (2010) Formulations with feruloyl glycerides and methods of preparations. US Patent 7:744,856
Zhao Z, Li P, Fan Q, Ya H (2019). Struct Chem 30:1707
Veselinović JB, Đorđević, BogdanovićIvana, VM, Morić I, Veselinović AM (2018) Struct Chem 29:541
Balaban AT (2019). Struct Chem 30:1129
Andrade-Ochoa S, Nevárez-Moorillόn GV, Sánchez-Torres LE, Villanueva-García M, Sánchez-Ramíez BE, Rodríguez-Valdez LM, Rivera-Chavira BE (2015). BMC Complement Altern Med 15:332
Pizzolitto RP, Jacquat AG, Usseglio VL, Achimón F, Cuillo AE, Zygadlo JA, Dambolena JS (2020). Food Control 108:106836
Crisan L, Borota A Bora A Pacureanu L (2019) Struct Chem 30:2311
Roy SM, Sharma BK, Roy DR (2019). Struct Chem 30:2379
Chavda J, Bhatt H (2019). Struct Chem 30:2093
Ambure P, Halder AL, Diaz HG, Cordeiro MNDS (2019). J Chem Inf Model 59:2538
Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, Dearden J, Gramatica P, Martin YC, Todeschini R, Consonni V, Kuz’min VE, Cramer R, Benigni R, Yang C, Rathman J, Terfloth L, Gasteiger J, Richard A, Tropsha A (2014). J Med Chem 57:4977
Topliss JG (1972). J Med Chem 15:1006
OECD (2007) Guidance document on the validation of (quantitative) structure-activity relationships [(Q)SAR] models, Paris
Yan J, Zhu WW, Kong B, Lu HB, Yun YH, Huang JH, Liang YZ (2014). Mol Inform 33:503
Ambure P, Aher RB, Gajewicz A, Puzyn T, Roy K (2015). Chemom Intell Lab Syst 147:1
Braga PC, Alfieri M, Culici M, Dal Sasso M (2007). Mycoses 50:502
Erturk MD, Sacan MT, Novic M, Minovski N (2012). J Mol Graph Model 38:90
Quiroga D, Becerra LD, Coy-Barrera E (2019). ACS Omega 4:13710
Duhan M, Singh R, Devi M, Sindhu J, Bhatia R, Kumar A, Kumar P (2019). J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2019.1704885
Acknowledgments
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture (USDA). USDA is an equal opportunity provider and employer and the provider. We are grateful for Prof. Paola Gramatica and Prof. Kanul Roy for the use of their software.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
All authors certify and assert that no animals or humans were used to obtain results reported in this research.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 63 kb)
Rights and permissions
About this article
Cite this article
Appell, M., Tu, YS., Compton, D.L. et al. Quantitative structure-activity relationship study for prediction of antifungal properties of phenolic compounds. Struct Chem 31, 1621–1630 (2020). https://doi.org/10.1007/s11224-020-01549-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-020-01549-1