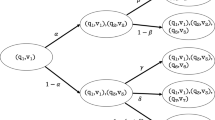
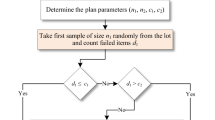
Abstract
Testing between hypotheses, when independent sampling is possible, is a well developed subject. In this paper, we propose hypothesis tests that are applicable when the samples are obtained using Markov chain Monte Carlo. These tests are useful when one is interested in deciding whether the expected value of a certain quantity is above or below a given threshold. We show non-asymptotic error bounds and bounds on the expected number of samples for three types of tests, a fixed sample size test, a sequential test with indifference region, and a sequential test without indifference region. Our tests can lead to significant savings in sample size. We illustrate our results on an example of Bayesian parameter inference involving an ODE model of a biochemical pathway.

Similar content being viewed by others
References
Bardenet, R., Doucet, A., Holmes, C.: Towards scaling up Markov chain Monte Carlo: an adaptive subsampling approach. Proceedings of the 31st International Conference on Machine Learning (ICML-14) (2014)
Gilks, W.R., Richardson, S., Spiegelhalter, D.J. (eds.): Markov Chain Monte Carlo in Practice. Interdisciplinary Statistics. Chapman & Hall, London (1996)
Gyori, B., Paulin, D.: Non-asymptotic confidence intervals for MCMC in practice. arXiv preprint (2014)
Kato, T.: Perturbation Theory for Linear Operators, vol. 132. Springer Science & Business Media, Heidelberg (1976)
Korattikara, A., Chen, Y., Welling, M.: Austerity in MCMC land: cutting the Metropolis–Hastings budget. Proceedings of the 31st International Conference on Machine Learning (ICML-14) (2014)
Klipp, E., Herwig, R., Kowald, A., Wierling, C., Lehrach, H.: Systems Biology in Practice: Concepts, Implementation and Application. Wiley-VCH, Weinheim (2005)
Lai, T.L.: Optimal stopping and sequential tests which minimize the maximum expected sample size. Ann. Stat. 1, 659–673 (1973)
Lai, T.L.: Nearly optimal sequential tests of composite hypotheses. Ann. Stat. 16(2), 856–886 (1988). doi:10.1214/aos/1176350840
Lawrence, N.D., Girolami, M., Rattray, M., Sanguinetti, G.: Learning and Inference in Computational Systems Biology. MIT Press, Cambridge (2009)
Legay, A., Delahaye, B., Bensalem, S.: Statistical Model Checking: An Overview. In: Barringer, H., Falcone, Y., Finkbeiner, B., Havelind, K., Lee, I., Pace, G., Rosu, G., Sokolsky, O., Tikkmann, N. (eds.) Runtime Verification, pp. 122–135. Springer, Heidelberg (2010)
Lehmann, E.L., Romano, J.P.: Testing Statistical Hypotheses, Springer Texts in Statistics, 3rd edn. Springer, New York (2005)
León, C.A., Perron, F.: Optimal Hoeffding bounds for discrete reversible Markov chains. Ann. Appl. Probab. 14(2), 958–970 (2004). doi:10.1214/105051604000000170
Miasojedow, B.: Hoeffding’s inequalities for geometrically ergodic Markov chains on general state space. Stat. Probab. Lett. 87, 115–120 (2014). doi:10.1016/j.spl.2014.01.013
Paulin, D.: Concentration inequalities for Markov chains by Marton couplings and spectral methods. Electron. J. Probab. (to appear)
Roberts, G.O., Rosenthal, J.S.: General state space Markov chains and MCMC algorithms. Probab. Surv. 1, 20–71 (2004). doi:10.1214/154957804100000024
Swameye, I., Müller, T., Jt, Timmer, Sandra, O., Klingmüller, U.: Identification of nucleocytoplasmic cycling as a remote sensor in cellular signaling by databased modeling. Proc. Natl. Acad. Sci. 100(3), 1028–1033 (2003)
Wald, A.: Sequential tests of statistical hypotheses. Ann. Math. Stat. 16(2), 117–186 (1945)
Acknowledgments
The authors thank the anonymous referees for their insightful comments. DP was supported by an MOE Singapore Academic Research Fund Tier 2 Grant “Approximate Computational Methods for High-Dimensional Systems”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Source code available at github.com/bgyori/mcmchyp.
Appendix
Appendix
1.1 Estimating the spectral gap
In this section, we propose a procedure to estimate the spectral gap of Markov chains. The procedure is motivated by the following lemma. In the statement of the lemma, we are going to use the scalar product
where \(\pi \) is the stationary distribution of a finite state Markov chain.
Lemma 1
Suppose that \(X_1,X_2,\ldots \) is a finite state Markov chain with spectral gap \(\gamma \), and transition kernel P. Suppose that \(f:\varOmega \rightarrow \mathbb {R}\) satisfies that \(\mathbb {E}_{\pi }(f)=0\), and that f is not orthogonal to the eigenspace corresponding to the eigenvector of P of the second largest absolute value (with respect to the scalar product \(\left<\cdot ,\cdot \right>_{\pi }\)). Let \(\rho _\eta (f):=\mathbb {E}_{\pi }(f(X_0)f(X_\eta ))\), then
Remark 1
A similar result can be shown for chains with general state spaces, but for notational simplicity we only consider finite state spaces. In practice, the condition of non-orthogonality is almost always satisfied.
Proof
We can write the eigenvalues of the operator P as \(1={\uplambda }_1\ge {\uplambda }_2\ge \ldots \ge {\uplambda }_n\), with \(n=|\varOmega |\), and corresponding right eigenvectors \(f_1,\ldots ,f_n\), which are orthonormal with respect to the scalar product \(\left<\cdot ,\cdot \right>_{\pi }\). It is clear that \(f_1=1\), and using the assumption \(\mathbb {E}_{\pi }(f)=0\), f can be decomposed as
for some constants \(\{c_i\}_{i=1}^{n}\). Using this decomposition, we have
and using the condition of non-orthogonality, we have
In practice, we cannot choose \(\eta \) to be infinity, in particular, there are some issues related to the variance of the estimator of \(\rho _{\eta }(f)\) that need to be taken in to account. Given burn-in time \(t_0\), and \(f(X_{t_{0}+1})\), \(\ldots , f(X_{t_0+n})\), we define the empirical mean as
and use the estimator
The typical range of dependence of the elements of the Markov chain is of the order of \(1/\gamma ^*\), so we expect the standard deviation of \(\hat{\rho }_{\eta }(f)/\mathrm {Var}_{\pi }(f)\) to be of the order of \(\frac{1}{\sqrt{n \gamma ^*}}\). Now if we want to use the estimator
the standard deviation of \(\rho _{\eta }(f)/\mathrm {Var}_{\pi }(f)\) needs to be much smaller than \((1-\gamma ^*)^{\eta }\).
Solving the equation \(\frac{1}{\sqrt{n \gamma ^*}}=(1-\gamma ^*)^{\eta }\) leads to \(\eta =\frac{\log (n\gamma ^*)}{2\log (1/(1-\gamma ^*))}\), so we propose the choice
for which the standard deviation of \(\rho _{\eta }(f)/\mathrm {Var}_{\pi }(f)\) is much smaller than \((1-\gamma ^*)^{\eta }\). A slight inconvenience of this choice is that it depends on the unknown parameter \(\gamma ^*\). A way to overcome this issue is by computing the value of \(\eta \) iteratively. Based on Lemma 1 and the above observation, we propose the following procedure.
-
1.
Choose some functions \(f_1,\ldots , f_m: \varOmega \rightarrow \mathbb {R}\) that together determine the location in the state space (for example, if \(\varOmega \subset \mathbb {R}^m\), then \(f_1,\ldots , f_m\) can be chosen as the coordinates).
-
2.
Run some initial amount of simulations \(X_1,\ldots , X_{t_0+n}\), and in every step, save the values \(f_1(X_i),\ldots , f_m(X_i)\).
-
3.
Compute \(\hat{\gamma }^*\) based on \(\eta =1\) and \(f=f_1, \ldots , f_m\). Denote the minimum of these values by \(\gamma ^*_{\min }(1)\). Compute \(\eta (1):=\frac{\log (n\gamma ^*)}{4\log (1/(1-\gamma ^*_{\min }(1)))}\).
-
4.
Inductively assume we have already computed \(\eta (i)\). Then compute \(\gamma ^*_{\min }(i+1)\) based on \(\eta (i)\). If \(\gamma ^*_{\min }(i+1)\ge \gamma ^*_{\min }(i)\), then stop, and let \(\hat{\gamma }^*:=\gamma ^*_{\min }(i)\). Otherwise compute \(\eta (i+1)\) and repeat this step.
-
5.
If the initial amount of simulations n satisfies that \(n>100/\hat{\gamma }^*\), accept the estimate, otherwise choose \(n=200/\hat{\gamma }^*\) and restart from Step 2.
The motivation for choosing \(f_1,\ldots , f_m\) in this way is that we use all the available information about the Markov chain, and thus the estimator is expected to be more accurate than if we would only use a subset of this information. The motivation for Step 5 is to ensure that we have sufficient initial data for the estimate.
An illustration of this iterative procedure based on our case study (see Sect. 4) is shown in Fig. 2. Here we use the components of the Markov chain’s state (the value of the model parameters \(k_1\) to \(k_4\)) for the estimation.
A histogram of estimated spectral gap values is shown in Figure 3 for the 1000 independent chains used in our case study.
1.2 Simulation details
Here we provide additional details on the JAK-STAT pathway model case study. The species in the model are listed in Table 1.
The ODE equations governing the model dynamics are as follows.
Here K is the number of delay variables, set to \(K=10\).
The observation model is defined as follows.
Here \(y_1\) represents total phosphorylated STAT and \(y_2\) represents total STAT in cytoplasm. Observations are available at 18 discrete time points up to 60 min. The data points, as well as standard deviations for each point are available in Swameye et al. (2003).
The parameter vector of the model is \(\varvec{\theta }=(k_1,k_2,\) \(k_3,k_4)\). We assume a uniform prior over a hypercube defined by a bounded interval for each parameter. The parameter ranges and the covariance matrix diagonal entries (\(\sigma _{\mathrm {MH}}\)) used to define the MCMC proposal distribution are provided in Table 2.
Rights and permissions
About this article
Cite this article
Gyori, B.M., Paulin, D. Hypothesis testing for Markov chain Monte Carlo. Stat Comput 26, 1281–1292 (2016). https://doi.org/10.1007/s11222-015-9594-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11222-015-9594-1